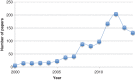
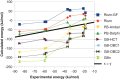
The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities
- PMID: 25835573
- PMCID: PMC4487606
- DOI: 10.1517/17460441.2015.1032936
The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities
Abstract
Introduction: The molecular mechanics energies combined with the Poisson-Boltzmann or generalized Born and surface area continuum solvation (MM/PBSA and MM/GBSA) methods are popular approaches to estimate the free energy of the binding of small ligands to biological macromolecules. They are typically based on molecular dynamics simulations of the receptor-ligand complex and are therefore intermediate in both accuracy and computational effort between empirical scoring and strict alchemical perturbation methods. They have been applied to a large number of systems with varying success.
Areas covered: The authors review the use of MM/PBSA and MM/GBSA methods to calculate ligand-binding affinities, with an emphasis on calibration, testing and validation, as well as attempts to improve the methods, rather than on specific applications.
Expert opinion: MM/PBSA and MM/GBSA are attractive approaches owing to their modular nature and that they do not require calculations on a training set. They have been used successfully to reproduce and rationalize experimental findings and to improve the results of virtual screening and docking. However, they contain several crude and questionable approximations, for example, the lack of conformational entropy and information about the number and free energy of water molecules in the binding site. Moreover, there are many variants of the method and their performance varies strongly with the tested system. Likewise, most attempts to ameliorate the methods with more accurate approaches, for example, quantum-mechanical calculations, polarizable force fields or improved solvation have deteriorated the results.
Keywords: drug design; electrostatics; entropy; free energy perturbation; linear interaction energy; non-polar solvation; solvation.
Figures
Similar articles
-
A head-to-head comparison of MM/PBSA and MM/GBSA in predicting binding affinities for the CB1 cannabinoid ligands.J Mol Model. 2024 Oct 31;30(11):390. doi: 10.1007/s00894-024-06189-4. J Mol Model. 2024. PMID: 39480515
-
Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking.Phys Chem Chem Phys. 2016 Aug 10;18(32):22129-39. doi: 10.1039/c6cp03670h. Phys Chem Chem Phys. 2016. PMID: 27444142
-
Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations.J Chem Inf Model. 2011 Jan 24;51(1):69-82. doi: 10.1021/ci100275a. Epub 2010 Nov 30. J Chem Inf Model. 2011. PMID: 21117705 Free PMC article.
-
Development and test of highly accurate endpoint free energy methods. 1: Evaluation of ABCG2 charge model on solvation free energy prediction and optimization of atom radii suitable for more accurate solvation free energy prediction by the PBSA method.J Comput Chem. 2023 May 30;44(14):1334-1346. doi: 10.1002/jcc.27089. Epub 2023 Feb 21. J Comput Chem. 2023. PMID: 36807356 Review.
-
Understanding the impact of binding free energy and kinetics calculations in modern drug discovery.Expert Opin Drug Discov. 2024 Jun;19(6):671-682. doi: 10.1080/17460441.2024.2349149. Epub 2024 May 9. Expert Opin Drug Discov. 2024. PMID: 38722032 Review.
Cited by
-
Family Malvaceae: a potential source of secondary metabolites with chemopreventive and anticancer activities supported with in silico pharmacokinetic and pharmacodynamic profiles.Front Pharmacol. 2024 Oct 16;15:1465055. doi: 10.3389/fphar.2024.1465055. eCollection 2024. Front Pharmacol. 2024. PMID: 39478959 Free PMC article.
-
Discovery of novel NLRP3 inhibitors based on machine learning and physical methods.BMC Chem. 2024 Oct 28;18(1):210. doi: 10.1186/s13065-024-01323-y. BMC Chem. 2024. PMID: 39468648 Free PMC article.
-
Computational Analysis of Residue-Specific Binding Free Energies of Androgen Receptor to Ligands.Front Mol Biosci. 2021 Mar 12;8:646524. doi: 10.3389/fmolb.2021.646524. eCollection 2021. Front Mol Biosci. 2021. PMID: 33778009 Free PMC article.
-
In Silico Study towards Repositioning of FDA-Approved Drug Candidates for Anticoronaviral Therapy: Molecular Docking, Molecular Dynamics and Binding Free Energy Calculations.Molecules. 2022 Sep 14;27(18):5988. doi: 10.3390/molecules27185988. Molecules. 2022. PMID: 36144718 Free PMC article.
-
Hybrid Caffeic Acid-Based DHFR Inhibitors as Novel Antimicrobial and Anticancer Agents.Antibiotics (Basel). 2024 May 23;13(6):479. doi: 10.3390/antibiotics13060479. Antibiotics (Basel). 2024. PMID: 38927146 Free PMC article.
References
-
- Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Ang Chem Int Ed. 2002;41:2644–76. - PubMed
-
Good review on computational approaches to ligand binding.
-
- Shirts MR, Mobley DL, Chodera JD. Alchemical free energy calculations: ready for prime time? Ann Rep Comput Chem. 2007;3:41–59.
-
- Homeyer N, Stoll F, Hillisch A, et al. Binding free energy calculations for lead optimization: assessment of their accuracy in an industrial drug design context. J Chem Theory Comput. 2014;10:3331–44. - PubMed
-
- Sham YY, Chu ZT, Tao H, et al. Examining methods for calculations of binding free energies: LRA, LIE, PDLD-LRA, and PDLD/S-LRA calculations of ligands binding to an HIV protease. Proteins. 2000;39:393–407. - PubMed
-
- Åqvist J, Medina C, Samuelsson JE. A new method for predicting binding affinity in computer-aided drug design. Proteins Eng. 1994;7:385–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical