Regulation of bacterial virulence by Csr (Rsm) systems
- PMID: 25833324
- PMCID: PMC4394879
- DOI: 10.1128/MMBR.00052-14
Regulation of bacterial virulence by Csr (Rsm) systems
Abstract
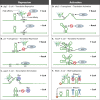
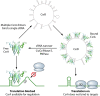
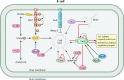
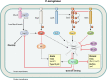
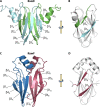
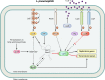
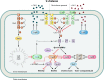
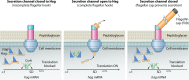
Most bacterial pathogens have the remarkable ability to flourish in the external environment and in specialized host niches. This ability requires their metabolism, physiology, and virulence factors to be responsive to changes in their surroundings. It is no surprise that the underlying genetic circuitry that supports this adaptability is multilayered and exceedingly complex. Studies over the past 2 decades have established that the CsrA/RsmA proteins, global regulators of posttranscriptional gene expression, play important roles in the expression of virulence factors of numerous proteobacterial pathogens. To accomplish these tasks, CsrA binds to the 5' untranslated and/or early coding regions of mRNAs and alters translation, mRNA turnover, and/or transcript elongation. CsrA activity is regulated by noncoding small RNAs (sRNAs) that contain multiple CsrA binding sites, which permit them to sequester multiple CsrA homodimers away from mRNA targets. Environmental cues sensed by two-component signal transduction systems and other regulatory factors govern the expression of the CsrA-binding sRNAs and, ultimately, the effects of CsrA on secretion systems, surface molecules and biofilm formation, quorum sensing, motility, pigmentation, siderophore production, and phagocytic avoidance. This review presents the workings of the Csr system, the paradigm shift that it generated for understanding posttranscriptional regulation, and its roles in virulence networks of animal and plant pathogens.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures







Similar articles
-
Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system.Curr Opin Microbiol. 2018 Feb;41:58-67. doi: 10.1016/j.mib.2017.11.022. Epub 2017 Dec 5. Curr Opin Microbiol. 2018. PMID: 29207313 Review.
-
Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems.Environ Microbiol. 2013 Feb;15(2):313-24. doi: 10.1111/j.1462-2920.2012.02794.x. Epub 2012 Jun 5. Environ Microbiol. 2013. PMID: 22672726 Free PMC article. Review.
-
Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB.Mol Microbiol. 1998 Sep;29(6):1321-30. doi: 10.1046/j.1365-2958.1998.01021.x. Mol Microbiol. 1998. PMID: 9781871 Review.
-
RsmV, a Small Noncoding Regulatory RNA in Pseudomonas aeruginosa That Sequesters RsmA and RsmF from Target mRNAs.J Bacteriol. 2018 Jul 25;200(16):e00277-18. doi: 10.1128/JB.00277-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29866805 Free PMC article.
-
Circuitry Linking the Catabolite Repression and Csr Global Regulatory Systems of Escherichia coli.J Bacteriol. 2016 Oct 7;198(21):3000-3015. doi: 10.1128/JB.00454-16. Print 2016 Nov 1. J Bacteriol. 2016. PMID: 27551019 Free PMC article.
Cited by
-
Emergence of a Competence-Reducing Filamentous Phage from the Genome of Acinetobacter baylyi ADP1.J Bacteriol. 2016 Nov 4;198(23):3209-3219. doi: 10.1128/JB.00424-16. Print 2016 Dec 1. J Bacteriol. 2016. PMID: 27645387 Free PMC article.
-
Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides.Chem Rev. 2022 Sep 28;122(18):14722-14814. doi: 10.1021/acs.chemrev.2c00210. Epub 2022 Sep 1. Chem Rev. 2022. PMID: 36049139 Free PMC article. Review.
-
Bacterial pathogenesis of plants: future challenges from a microbial perspective: Challenges in Bacterial Molecular Plant Pathology.Mol Plant Pathol. 2016 Oct;17(8):1298-313. doi: 10.1111/mpp.12427. Epub 2016 Aug 4. Mol Plant Pathol. 2016. PMID: 27170435 Free PMC article. Review.
-
Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis.Proc Natl Acad Sci U S A. 2016 Jun 28;113(26):7237-42. doi: 10.1073/pnas.1523004113. Epub 2016 Jun 13. Proc Natl Acad Sci U S A. 2016. PMID: 27298343 Free PMC article.
-
Discrimination and Integration of Stress Signals by Pathogenic Bacteria.Cell Host Microbe. 2016 Aug 10;20(2):144-153. doi: 10.1016/j.chom.2016.07.010. Cell Host Microbe. 2016. PMID: 27512902 Free PMC article. Review.
References
-
- Carmichael GG, Weber K, Niveleau A, Wahba AJ. 1975. The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J Biol Chem 250:3607–3612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

