Diverse selective regimes shape genetic diversity at ADAR genes and at their coding targets
- PMID: 25826567
- PMCID: PMC4615741
- DOI: 10.1080/15476286.2015.1017215
Diverse selective regimes shape genetic diversity at ADAR genes and at their coding targets
Abstract
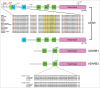
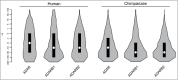
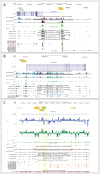
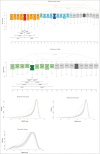
A-to-I RNA editing operated by ADAR enzymes is extremely common in mammals. Several editing events in coding regions have pivotal physiological roles and affect protein sequence (recoding events) or function. We analyzed the evolutionary history of the 3 ADAR family genes and of their coding targets. Evolutionary analysis indicated that ADAR evolved adaptively in primates, with the strongest selection in the unique N-terminal domain of the interferon-inducible isoform. Positively selected residues in the human lineage were also detected in the ADAR deaminase domain and in the RNA binding domains of ADARB1 and ADARB2. During the recent history of human populations distinct variants in the 3 genes increased in frequency as a result of local selective pressures. Most selected variants are located within regulatory regions and some are in linkage disequilibrium with eQTLs in monocytes. Finally, analysis of conservation scores of coding editing sites indicated that editing events are counter-selected within regions that are poorly tolerant to change. Nevertheless, a minority of recoding events occurs at highly conserved positions and possibly represents the functional fraction. These events are enriched in pathways related to HIV-1 infection and to epidermis/hair development. Thus, both ADAR genes and their targets evolved under variable selective regimes, including purifying and positive selection. Pressures related to immune response likely represented major drivers of evolution for ADAR genes. As for their coding targets, we suggest that most editing events are slightly deleterious, although a minority may be beneficial and contribute to antiviral response and skin homeostasis.
Keywords: 1000G,1000 Genomes Pilot Project; A to I, adenosine to inosine; A-to-I editing; ADAR; ADAR editing sites; AGS, Aicardi-Goutières Syndrome; BEB, Bayes Empirical Bayes; BS-REL, branch site-random effects likelihood; CEU, Europeans; CHBJPT, Chinese plus Japanese; DAF, derived allele frequency; DIND, Derived Intra-allelic Nucleotide Diversity; DSH, dyschromatosis symmetrica hereditaria; FDR, false discovery rate; GARD, Genetic Algorithm Recombination Detection; GERP Genomic Evolutionary Rate Profiling; IFN, Interferon; LD, linkage disequilibrium; LRT, likelihood ratio test; MAF, minor allele frequency; MEME, Mixed Effects Model of Evolution; RBD, dsRNA binding domain; SLAC, single-likelihood ancestor counting; YRI, Yoruba; eQTL, Expression quantitative trait loci; evolutionary analysis; iHS, Integrated Haplotype Score; positive selection.
Figures
Similar articles
-
Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase.J Biol Chem. 1997 Feb 14;272(7):4419-28. doi: 10.1074/jbc.272.7.4419. J Biol Chem. 1997. PMID: 9020165
-
What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs.Open Biol. 2020 Jul;10(7):200085. doi: 10.1098/rsob.200085. Epub 2020 Jul 1. Open Biol. 2020. PMID: 32603639 Free PMC article. Review.
-
The role of RNA editing enzyme ADAR1 in human disease.Wiley Interdiscip Rev RNA. 2022 Jan;13(1):e1665. doi: 10.1002/wrna.1665. Epub 2021 Jun 8. Wiley Interdiscip Rev RNA. 2022. PMID: 34105255 Free PMC article. Review.
-
Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1.Biol Reprod. 2017 Jan 1;96(1):244-253. doi: 10.1095/biolreprod.116.145151. Biol Reprod. 2017. PMID: 28395340 Free PMC article.
-
ADAR RNA editing below the backbone.RNA. 2017 Sep;23(9):1317-1328. doi: 10.1261/rna.060921.117. Epub 2017 May 30. RNA. 2017. PMID: 28559490 Free PMC article. Review.
Cited by
-
ALU non-B-DNA conformations, flipons, binary codes and evolution.R Soc Open Sci. 2020 Jun 3;7(6):200222. doi: 10.1098/rsos.200222. eCollection 2020 Jun. R Soc Open Sci. 2020. PMID: 32742689 Free PMC article. Review.
-
ADAR1 Prevents R-loop Accumulation-Driven ATR Pathway Activation in Ovarian Cancer.J Cancer. 2022 Apr 24;13(8):2397-2412. doi: 10.7150/jca.72108. eCollection 2022. J Cancer. 2022. PMID: 35711824 Free PMC article.
-
Mapping the dsRNA World.Cold Spring Harb Perspect Biol. 2019 Mar 1;11(3):a035352. doi: 10.1101/cshperspect.a035352. Cold Spring Harb Perspect Biol. 2019. PMID: 30824577 Free PMC article. Review.
-
A novel missense mutation of ADAR1 gene in a Chinese family leading to dyschromatosis symmetrica hereditaria and literature review.J Genet. 2017 Dec;96(6):1021-1026. doi: 10.1007/s12041-017-0873-9. J Genet. 2017. PMID: 29321362
-
Regulation and functional consequences of mGlu4 RNA editing.RNA. 2021 Oct;27(10):1220-1240. doi: 10.1261/rna.078729.121. Epub 2021 Jul 8. RNA. 2021. PMID: 34244459 Free PMC article.
References
-
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 2010; 79:321-349; PMID:20192758; http://dx.doi.org/10.1146/annurev-biochem-060208-105251 - DOI - PMC - PubMed
-
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. . A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 2014; 24:365-376; PMID:24347612; http://dx.doi.org/10.1101/gr.164749.113 - DOI - PMC - PubMed
-
- Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol 2012; 13:252; PMID:23273215; http://dx.doi.org/10.1186/gb-2012-13-12-252 - DOI - PMC - PubMed
-
- Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med 2013; 5:105; PMID:24289319; http://dx.doi.org/10.1186/gm508 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
