Mechanistic insights on the Dicer-independent AGO2-mediated processing of AgoshRNAs
- PMID: 25826416
- PMCID: PMC4615756
- DOI: 10.1080/15476286.2015.1017204
Mechanistic insights on the Dicer-independent AGO2-mediated processing of AgoshRNAs
Abstract
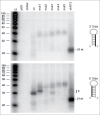
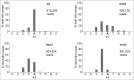
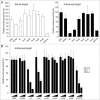
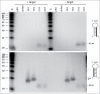
Short hairpin RNAs (shRNAs) are widely used for gene knockdown by inducing the RNA interference (RNAi) mechanism, both for research and therapeutic purposes. The shRNA precursor is processed by the RNase III-like enzyme Dicer into biologically active small interfering RNA (siRNA). This effector molecule subsequently targets a complementary mRNA for destruction via the Argonaute 2 (AGO2) complex. The cellular role of Dicer concerns the processing of pre-miRNAs into mature microRNA (miRNA). Recently, a non-canonical pathway was reported for the biogenesis of miR-451, which bypasses Dicer and is processed instead by the slicer activity of AGO2, followed by the regular AGO2-mediated mRNA targeting step. Interestingly, shRNA designs that are characterized by a relatively short basepaired stem also bypass Dicer to be processed by AGO2. We named this design AgoshRNA as these molecules depend on AGO2 both for processing and silencing activity. In this study, we investigated diverse mechanistic aspects of this new class of AgoshRNA molecules. We probed the requirements for AGO2-mediated processing of AgoshRNAs by modification of the proposed cleavage site in the hairpin. We demonstrate by deep sequencing that AGO2-processed AgoshRNAs produce RNA effector molecules with more discrete ends than the products of the regular shRNA design. Furthermore, we tested whether trimming and tailing occurs upon AGO2-mediated processing of AgoshRNAs, similar to what has been described for miR-451. Finally, we tested the prediction that AgoshRNA activity, unlike that of regular shRNAs, is maintained in Dicer-deficient cell types. These mechanistic insights could aid in the design of optimised AgoshRNA tools and therapeutics.
Keywords: AgoshRNA; Argonaute 2; Dicer; RNA processing; shRNA.
Figures



Similar articles
-
Influence of the loop size and nucleotide composition on AgoshRNA biogenesis and activity.RNA Biol. 2017 Nov 2;14(11):1559-1569. doi: 10.1080/15476286.2017.1328349. Epub 2017 Nov 3. RNA Biol. 2017. PMID: 28569591 Free PMC article.
-
Analysis of AgoshRNA maturation and loading into Ago2.PLoS One. 2017 Aug 15;12(8):e0183269. doi: 10.1371/journal.pone.0183269. eCollection 2017. PLoS One. 2017. PMID: 28809941 Free PMC article.
-
Towards Antiviral shRNAs Based on the AgoshRNA Design.PLoS One. 2015 Jun 18;10(6):e0128618. doi: 10.1371/journal.pone.0128618. eCollection 2015. PLoS One. 2015. PMID: 26087209 Free PMC article.
-
Dicer-independent processing of small RNA duplexes: mechanistic insights and applications.Nucleic Acids Res. 2017 Oct 13;45(18):10369-10379. doi: 10.1093/nar/gkx779. Nucleic Acids Res. 2017. PMID: 28977573 Free PMC article. Review.
-
Biogenesis and Function of Ago-Associated RNAs.Trends Genet. 2017 Mar;33(3):208-219. doi: 10.1016/j.tig.2017.01.003. Epub 2017 Feb 4. Trends Genet. 2017. PMID: 28174021 Review.
Cited by
-
MicroRNAs Are Involved in the Development of Morphine-Induced Analgesic Tolerance and Regulate Functionally Relevant Changes in Serpini1.Front Mol Neurosci. 2016 Mar 24;9:20. doi: 10.3389/fnmol.2016.00020. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27047334 Free PMC article.
-
SmithRNAs: Could Mitochondria "Bend" Nuclear Regulation?Mol Biol Evol. 2017 Aug 1;34(8):1960-1973. doi: 10.1093/molbev/msx140. Mol Biol Evol. 2017. PMID: 28444389 Free PMC article.
-
VEGFA-targeting miR-agshRNAs combine efficacy with specificity and safety for retinal gene therapy.Mol Ther Nucleic Acids. 2022 Feb 28;28:58-76. doi: 10.1016/j.omtn.2022.02.019. eCollection 2022 Jun 14. Mol Ther Nucleic Acids. 2022. PMID: 35356684 Free PMC article.
-
Micromanaging aerobic respiration and glycolysis in cancer cells.Mol Metab. 2019 May;23:98-126. doi: 10.1016/j.molmet.2019.01.014. Epub 2019 Feb 6. Mol Metab. 2019. PMID: 30837197 Free PMC article. Review.
-
Blocking sense-strand activity improves potency, safety and specificity of anti-hepatitis B virus short hairpin RNA.EMBO Mol Med. 2016 Sep 1;8(9):1082-98. doi: 10.15252/emmm.201506172. Print 2016 Sep. EMBO Mol Med. 2016. PMID: 27473329 Free PMC article.
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; http://dx.doi.org/10.1038/35888 - DOI - PubMed
-
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990; 2:279-89; PMID:12354959; http://dx.doi.org/10.1105/tpc.2.4.279 - DOI - PMC - PubMed
-
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/10.1016/j.cell.2009.01.002 - DOI - PMC - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004; 432:231-5; PMID:15531879; http://dx.doi.org/10.1038/nature03049 - DOI - PubMed
-
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432:235-40; PMID:15531877 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
