Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses
- PMID: 25818028
- PMCID: PMC4426963
- DOI: 10.1016/j.virol.2015.03.001
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses
Abstract
Plus strand RNA viruses that replicate in the cytoplasm face challenges in supporting the numerous biosynthetic functions required for replication and propagation. Most of these viruses are genetically simple and rely heavily on co-opting cellular proteins, particularly cellular RNA-binding proteins, into new roles for support of virus infection at the level of virus-specific translation, and building RNA replication complexes. In the course of infectious cycles many nuclear-cytoplasmic shuttling proteins of mostly nuclear distribution are detained in the cytoplasm by viruses and re-purposed for their own gain. Many mammalian viruses hijack a common group of the same factors. This review summarizes recent gains in our knowledge of how cytoplasmic RNA viruses use these co-opted host nuclear factors in new functional roles supporting virus translation and virus RNA replication and common themes employed between different virus groups.
Keywords: Coronavirus; Enterovirus; Flavivirus; G3BP1; Hepatitis C virus; La protein; NSAP; Norovirus; PCBP2; PTB; Poliovirus; RNA helicase A; RNA virus; SRp20; Tia1/TIAR; UNR; hnRNP A1; hnRNP C; hnRNP K; hnRNP M.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
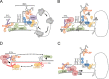
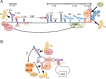
Similar articles
-
Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1.J Virol. 2013 Nov;87(21):11371-87. doi: 10.1128/JVI.03433-12. Epub 2013 Aug 14. J Virol. 2013. PMID: 23946460 Free PMC article.
-
The dependence of viral RNA replication on co-opted host factors.Nat Rev Microbiol. 2011 Dec 19;10(2):137-49. doi: 10.1038/nrmicro2692. Nat Rev Microbiol. 2011. PMID: 22183253 Free PMC article. Review.
-
Diverse roles of host RNA binding proteins in RNA virus replication.RNA Biol. 2011 Mar-Apr;8(2):305-15. doi: 10.4161/rna.8.2.15391. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21505273 Free PMC article. Review.
-
The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication.Virus Res. 2003 Sep;95(1-2):13-22. doi: 10.1016/s0168-1702(03)00160-6. Virus Res. 2003. PMID: 12921992 Free PMC article. Review.
-
Direct and Indirect Effects on Viral Translation and RNA Replication Are Required for AUF1 Restriction of Enterovirus Infections in Human Cells.mBio. 2018 Sep 4;9(5):e01669-18. doi: 10.1128/mBio.01669-18. mBio. 2018. PMID: 30181254 Free PMC article.
Cited by
-
Nucleolin Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus by Supporting the Assembly of Translation Initiation Complexes.J Virol. 2021 Jun 10;95(13):e0023821. doi: 10.1128/JVI.00238-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853964 Free PMC article.
-
The multifarious roles of heterogeneous ribonucleoprotein A1 in viral infections.Rev Med Virol. 2020 Mar;30(2):e2097. doi: 10.1002/rmv.2097. Epub 2020 Jan 27. Rev Med Virol. 2020. PMID: 31989716 Free PMC article. Review.
-
Seneca Valley virus 3Cpro degrades heterogeneous nuclear ribonucleoprotein A1 to facilitate viral replication.Virulence. 2021 Dec;12(1):3125-3136. doi: 10.1080/21505594.2021.2014681. Virulence. 2021. PMID: 34923914 Free PMC article.
-
Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases.Front Microbiol. 2018 Jun 19;9:1324. doi: 10.3389/fmicb.2018.01324. eCollection 2018. Front Microbiol. 2018. PMID: 29971060 Free PMC article.
-
Selective Removal of FG Repeat Domains from the Nuclear Pore Complex by Enterovirus 2A(pro).J Virol. 2015 Nov;89(21):11069-79. doi: 10.1128/JVI.00956-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311873 Free PMC article.
References
-
- Agis-Juárez R.A., Galván I., Medina F., Daikoku T., Padmanabhan R., Ludert J.E., del Angel R.M. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J. Gen.Virol. 2009;90:2893–2901. doi: 10.1099/vir.0.013433-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

