PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway
- PMID: 25817994
- PMCID: PMC4432305
- DOI: 10.1074/jbc.M115.646521
PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway
Abstract
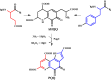
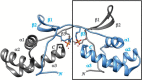
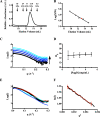
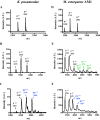
Pyrroloquinoline quinone (PQQ) is a product of a ribosomally synthesized and post-translationally modified pathway consisting of five conserved genes, pqqA-E. PqqE is a radical S-adenosylmethionine (RS) protein with a C-terminal SPASM domain, and is proposed to catalyze the formation of a carbon-carbon bond between the glutamate and tyrosine side chains of the peptide substrate PqqA. PqqD is a 10-kDa protein with an unknown function, but is essential for PQQ production. Recently, in Klebsiella pneumoniae (Kp), PqqD and PqqE were shown to interact; however, the stoichiometry and KD were not obtained. Here, we show that the PqqE and PqqD interaction transcends species, also occurring in Methylobacterium extorquens AM1 (Me). The stoichiometry of the MePqqD and MePqqE interaction is 1:1 and the KD, determined by surface plasmon resonance spectroscopy (SPR), was found to be ∼12 μm. Moreover, using SPR and isothermal calorimetry techniques, we establish for the first time that MePqqD binds MePqqA tightly (KD ∼200 nm). The formation of a ternary MePqqA-D-E complex was captured by native mass spectrometry and the KD for the MePqqAD-MePqqE interaction was found to be ∼5 μm. Finally, using a bioinformatic analysis, we found that PqqD orthologues are associated with the RS-SPASM family of proteins (subtilosin, pyrroloquinoline quinone, anaerobic sulfatase maturating enzyme, and mycofactocin), all of which modify either peptides or proteins. In conclusion, we propose that PqqD is a novel peptide chaperone and that PqqD orthologues may play a similar role in peptide modification pathways that use an RS-SPASM protein.
Keywords: PqqD; PqqE; peptide biosynthesis; peptide interaction; protein complex; quinone; small-angle x-ray scattering (SAXS).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
(1)H, (13)C, and (15)N resonance assignments and secondary structure information for Methylobacterium extorquens PqqD and the complex of PqqD with PqqA.Biomol NMR Assign. 2016 Oct;10(2):385-9. doi: 10.1007/s12104-016-9705-8. Epub 2016 Sep 16. Biomol NMR Assign. 2016. PMID: 27638737 Free PMC article.
-
Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD.J Biol Chem. 2016 Apr 22;291(17):8877-84. doi: 10.1074/jbc.C115.699918. Epub 2016 Mar 8. J Biol Chem. 2016. PMID: 26961875 Free PMC article.
-
Nuclear Magnetic Resonance Structure and Binding Studies of PqqD, a Chaperone Required in the Biosynthesis of the Bacterial Dehydrogenase Cofactor Pyrroloquinoline Quinone.Biochemistry. 2017 May 30;56(21):2735-2746. doi: 10.1021/acs.biochem.7b00247. Epub 2017 May 12. Biochemistry. 2017. PMID: 28481092 Free PMC article.
-
Biogenesis of the peptide-derived redox cofactor pyrroloquinoline quinone.Curr Opin Chem Biol. 2020 Dec;59:93-103. doi: 10.1016/j.cbpa.2020.05.001. Epub 2020 Jul 27. Curr Opin Chem Biol. 2020. PMID: 32731194 Free PMC article. Review.
-
Occurrence, function, and biosynthesis of mycofactocin.Appl Microbiol Biotechnol. 2019 Apr;103(7):2903-2912. doi: 10.1007/s00253-019-09684-4. Epub 2019 Feb 18. Appl Microbiol Biotechnol. 2019. PMID: 30778644 Free PMC article. Review.
Cited by
-
(1)H, (13)C, and (15)N resonance assignments and secondary structure information for Methylobacterium extorquens PqqD and the complex of PqqD with PqqA.Biomol NMR Assign. 2016 Oct;10(2):385-9. doi: 10.1007/s12104-016-9705-8. Epub 2016 Sep 16. Biomol NMR Assign. 2016. PMID: 27638737 Free PMC article.
-
Biosynthetic Timing and Substrate Specificity for the Thiopeptide Thiomuracin.J Am Chem Soc. 2016 Dec 7;138(48):15511-15514. doi: 10.1021/jacs.6b08987. Epub 2016 Oct 13. J Am Chem Soc. 2016. PMID: 27700071 Free PMC article.
-
RaS-RiPPs in Streptococci and the Human Microbiome.ACS Bio Med Chem Au. 2022 Aug 17;2(4):328-339. doi: 10.1021/acsbiomedchemau.2c00004. Epub 2022 Mar 21. ACS Bio Med Chem Au. 2022. PMID: 35996476 Free PMC article. Review.
-
Hydrogen-Deuterium Exchange Mass Spectrometry Identifies Local and Long-Distance Interactions within the Multicomponent Radical SAM Enzyme, PqqE.ACS Cent Sci. 2024 Jan 17;10(2):251-263. doi: 10.1021/acscentsci.3c01023. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435514 Free PMC article.
-
Structural Properties and Catalytic Implications of the SPASM Domain Iron-Sulfur Clusters in Methylorubrum extorquens PqqE.J Am Chem Soc. 2020 Jul 22;142(29):12620-12634. doi: 10.1021/jacs.0c02044. Epub 2020 Jul 9. J Am Chem Soc. 2020. PMID: 32643933 Free PMC article.
References
-
- Babasaki K., Takao T., Shimonishi Y., Kurahashi K. (1985) Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98, 585–603 - PubMed
-
- Duquesne S., Destoumieux-Garzón D. (2007) Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 24, 708–734 - PubMed
-
- Willey J. M., van der Donk W. A. (2007) Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501 - PubMed
-
- Okada M., Sato I., Cho S. J., Iwata H., Nishio T., Dubnau D., Sakagami Y. (2005) Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1, 23–24 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

