Decoding mechanisms by which silent codon changes influence protein biogenesis and function
- PMID: 25817479
- PMCID: PMC4461553
- DOI: 10.1016/j.biocel.2015.03.011
Decoding mechanisms by which silent codon changes influence protein biogenesis and function
Abstract
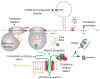
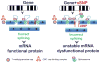
Scope: Synonymous codon usage has been a focus of investigation since the discovery of the genetic code and its redundancy. The occurrences of synonymous codons vary between species and within genes of the same genome, known as codon usage bias. Today, bioinformatics and experimental data allow us to compose a global view of the mechanisms by which the redundancy of the genetic code contributes to the complexity of biological systems from affecting survival in prokaryotes, to fine tuning the structure and function of proteins in higher eukaryotes. Studies analyzing the consequences of synonymous codon changes in different organisms have revealed that they impact nucleic acid stability, protein levels, structure and function without altering amino acid sequence. As such, synonymous mutations inevitably contribute to the pathogenesis of complex human diseases. Yet, fundamental questions remain unresolved regarding the impact of silent mutations in human disorders. In the present review we describe developments in this area concentrating on mechanisms by which synonymous mutations may affect protein function and human health.
Purpose: This synopsis illustrates the significance of synonymous mutations in disease pathogenesis. We review the different steps of gene expression affected by silent mutations, and assess the benefits and possible harmful effects of codon optimization applied in the development of therapeutic biologics.
Physiological and medical relevance: Understanding mechanisms by which synonymous mutations contribute to complex diseases such as cancer, neurodegeneration and genetic disorders, including the limitations of codon-optimized biologics, provides insight concerning interpretation of silent variants and future molecular therapies.
Keywords: Codon usage bias (CUB); Protein folding; Translation dynamics; mRNA structure; sSNP.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
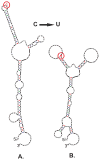
Similar articles
-
Bicodon bias can determine the role of synonymous SNPs in human diseases.BMC Genomics. 2017 Mar 13;18(1):227. doi: 10.1186/s12864-017-3609-6. BMC Genomics. 2017. PMID: 28288557 Free PMC article.
-
Molecular Mechanisms and the Significance of Synonymous Mutations.Biomolecules. 2024 Jan 20;14(1):132. doi: 10.3390/biom14010132. Biomolecules. 2024. PMID: 38275761 Free PMC article. Review.
-
Synonymous but Not Silent: The Codon Usage Code for Gene Expression and Protein Folding.Annu Rev Biochem. 2021 Jun 20;90:375-401. doi: 10.1146/annurev-biochem-071320-112701. Epub 2021 Jan 13. Annu Rev Biochem. 2021. PMID: 33441035 Free PMC article.
-
Codon-Resolution Analysis Reveals a Direct and Context-Dependent Impact of Individual Synonymous Mutations on mRNA Level.Mol Biol Evol. 2017 Nov 1;34(11):2944-2958. doi: 10.1093/molbev/msx229. Mol Biol Evol. 2017. PMID: 28961875 Free PMC article.
-
Roles for Synonymous Codon Usage in Protein Biogenesis.Annu Rev Biophys. 2015;44:143-66. doi: 10.1146/annurev-biophys-060414-034333. Epub 2015 Feb 26. Annu Rev Biophys. 2015. PMID: 25747594 Review.
Cited by
-
Novel compound heterozygous synonymous and missense variants in the MYO7A gene identified by next-generation sequencing in a Chinese family with nonsyndromic hearing loss.J Clin Lab Anal. 2022 Nov;36(11):e24708. doi: 10.1002/jcla.24708. Epub 2022 Sep 26. J Clin Lab Anal. 2022. PMID: 36164746 Free PMC article.
-
Mechanistic Approaches to Improve Correction of the Most Common Disease-Causing Mutation in Cystic Fibrosis.PLoS One. 2016 May 23;11(5):e0155882. doi: 10.1371/journal.pone.0155882. eCollection 2016. PLoS One. 2016. PMID: 27214033 Free PMC article.
-
A synonymous germline variant in a gene encoding a cell adhesion molecule is associated with cutaneous mast cell tumour development in Labrador and Golden Retrievers.PLoS Genet. 2019 Mar 22;15(3):e1007967. doi: 10.1371/journal.pgen.1007967. eCollection 2019 Mar. PLoS Genet. 2019. PMID: 30901340 Free PMC article.
-
The roles of RNA processing in translating genotype to phenotype.Nat Rev Mol Cell Biol. 2017 Feb;18(2):102-114. doi: 10.1038/nrm.2016.139. Epub 2016 Nov 16. Nat Rev Mol Cell Biol. 2017. PMID: 27847391 Free PMC article. Review.
-
Bicodon bias can determine the role of synonymous SNPs in human diseases.BMC Genomics. 2017 Mar 13;18(1):227. doi: 10.1186/s12864-017-3609-6. BMC Genomics. 2017. PMID: 28288557 Free PMC article.
References
-
- ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med - PubMed
-
- ADZHUBEI AA, ADZHUBEI IA, KRASHENINNIKOV IA, NEIDLE S. Non-random usage of ‘degenerate’ codons is related to protein three-dimensional structure. FEBS Lett. 1996;399:78–82. - PubMed
-
- ALATTIA JR, MATASCI M, DIMITROV M, AESCHBACH L, BALASUBRAMANIAN S, HACKER DL, WURM FM, FRAERING PC. Highly efficient production of the Alzheimer’s gamma-secretase integral membrane protease complex by a multi-gene stable integration approach. Biotechnol Bioeng. 2013;110:1995–2005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

