Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota
- PMID: 25811034
- PMCID: PMC4355334
- DOI: 10.1155/2015/361604
Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota
Abstract
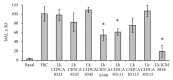
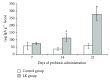
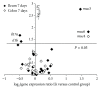
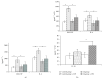
The evaluation of the impact of probiotics on host health could help to understand how they can be used in the prevention of diseases. On the basis of our previous studies and in vitro assays on PBMC and Caco-2 ccl20:luc reporter system presented in this work, the strain Lactobacillus kefiri CIDCA 8348 was selected and administrated to healthy Swiss mice daily for 21 days. The probiotic treatment increased IgA in feces and reduced expression of proinflammatory mediators in Peyer Patches and mesenteric lymph nodes, where it also increased IL-10. In ileum IL-10, CXCL-1 and mucin 6 genes were upregulated; meanwhile in colon mucin 4 was induced whereas IFN-γ, GM-CSF, and IL-1β genes were downregulated. Moreover, ileum and colon explants showed the anti-inflammatory effect of L. kefiri since the LPS-induced increment of IL-6 and GM-CSF levels in control mice was significantly attenuated in L. kefiri treated mice. Regarding fecal microbiota, DGGE profiles allowed differentiation of experimental groups in two separated clusters. Quantitative PCR analysis of different bacterial groups revealed only significant changes in Lactobacillus population. In conclusion, L. kefiri is a good candidate to be used in gut inflammatory disorders.
Figures

Similar articles
-
The Probiotic Compound VSL#3 Modulates Mucosal, Peripheral, and Systemic Immunity Following Murine Broad-Spectrum Antibiotic Treatment.Front Cell Infect Microbiol. 2017 May 5;7:167. doi: 10.3389/fcimb.2017.00167. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28529928 Free PMC article.
-
Adhesion properties of potentially probiotic Lactobacillus kefiri to gastrointestinal mucus.J Dairy Res. 2014 Feb;81(1):16-23. doi: 10.1017/S0022029913000526. Epub 2013 Oct 29. J Dairy Res. 2014. PMID: 24168928
-
Mucosal immunomodulation by the non-bacterial fraction of milk fermented by Lactobacillus helveticus R389.Int J Food Microbiol. 2007 Apr 10;115(2):180-6. doi: 10.1016/j.ijfoodmicro.2006.10.020. Epub 2006 Dec 20. Int J Food Microbiol. 2007. PMID: 17184869
-
Re-thinking the functions of IgA(+) plasma cells.Gut Microbes. 2014;5(5):652-62. doi: 10.4161/19490976.2014.969977. Gut Microbes. 2014. PMID: 25483334 Free PMC article. Review.
-
Intestinal Flora and Disease Mutually Shape the Regional Immune System in the Intestinal Tract.Front Immunol. 2020 Apr 3;11:575. doi: 10.3389/fimmu.2020.00575. eCollection 2020. Front Immunol. 2020. PMID: 32318067 Free PMC article. Review.
Cited by
-
The regulatory function of Blastocystis spp. on the immune inflammatory response in the gut microbiome.Front Cell Infect Microbiol. 2022 Aug 31;12:967724. doi: 10.3389/fcimb.2022.967724. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36118018 Free PMC article. Review.
-
Beneficial health effects of polyphenols metabolized by fermentation.Food Sci Biotechnol. 2022 Jun 27;31(8):1027-1040. doi: 10.1007/s10068-022-01112-0. eCollection 2022 Jul. Food Sci Biotechnol. 2022. PMID: 35873377 Free PMC article. Review.
-
Composition of the maternal gastrointestinal microbiome as a predictor of neonatal birth weight.Pediatr Res. 2023 Sep;94(3):1158-1165. doi: 10.1038/s41390-023-02584-4. Epub 2023 Apr 7. Pediatr Res. 2023. PMID: 37029236
-
Effect of Lactobacillus kefiri, in Conjunction with PENS T6 and a Hypocaloric Diet, on Weight Loss, Hypertension and Laboratory Glycemic and Lipid Profile.Nutrients. 2023 Oct 26;15(21):4549. doi: 10.3390/nu15214549. Nutrients. 2023. PMID: 37960202 Free PMC article.
-
Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir.mSystems. 2016 Oct 4;1(5):e00052-16. doi: 10.1128/mSystems.00052-16. eCollection 2016 Sep-Oct. mSystems. 2016. PMID: 27822552 Free PMC article.
References
-
- Delcenserie V., Martel D., Lamoureux M., Amiot J., Boutin Y., Roy D. Immunomodulatory effects of probiotics in the intestinal tract. Current Issues in Molecular Biology. 2008;10(1):37–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

