Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production
- PMID: 25810521
- PMCID: PMC4389594
- DOI: 10.1523/JNEUROSCI.3081-14.2015
Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production
Erratum in
- J Neurosci. 2015 Jun 3;35(22):8686. Adelita, Tai [corrected to Adelita, Tais]
Abstract
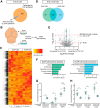
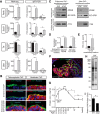
A sheet of choroid plexus epithelial cells extends into each cerebral ventricle and secretes signaling factors into the CSF. To evaluate whether differences in the CSF proteome across ventricles arise, in part, from regional differences in choroid plexus gene expression, we defined the transcriptome of lateral ventricle (telencephalic) versus fourth ventricle (hindbrain) choroid plexus. We find that positional identities of mouse, macaque, and human choroid plexi derive from gene expression domains that parallel their axial tissues of origin. We then show that molecular heterogeneity between telencephalic and hindbrain choroid plexi contributes to region-specific, age-dependent protein secretion in vitro. Transcriptome analysis of FACS-purified choroid plexus epithelial cells also predicts their cell-type-specific secretome. Spatial domains with distinct protein expression profiles were observed within each choroid plexus. We propose that regional differences between choroid plexi contribute to dynamic signaling gradients across the mammalian cerebroventricular system.
Keywords: cerebrospinal fluid; choroid plexus; next-generation sequencing.
Copyright © 2015 the authors 0270-6474/15/354903-14$15.00/0.
Figures
Similar articles
-
Transporter-mediated L-glutamate elimination from cerebrospinal fluid: possible involvement of excitatory amino acid transporters expressed in ependymal cells and choroid plexus epithelial cells.Fluids Barriers CNS. 2015 Apr 29;12:11. doi: 10.1186/s12987-015-0006-x. Fluids Barriers CNS. 2015. PMID: 25925580 Free PMC article.
-
Cellular transfer of macromolecules across the developing choroid plexus of Monodelphis domestica.Eur J Neurosci. 2009 Jan;29(2):253-66. doi: 10.1111/j.1460-9568.2008.06571.x. Eur J Neurosci. 2009. PMID: 19200232
-
microRNA-449 is a putative regulator of choroid plexus development and function.Brain Res. 2009 Jan 23;1250:20-6. doi: 10.1016/j.brainres.2008.11.020. Epub 2008 Nov 18. Brain Res. 2009. PMID: 19056356
-
Usefulness and limitation of primary cultured porcine choroid plexus epithelial cells as an in vitro model to study drug transport at the blood-CSF barrier.Adv Drug Deliv Rev. 2004 Oct 14;56(12):1859-73. doi: 10.1016/j.addr.2004.07.012. Adv Drug Deliv Rev. 2004. PMID: 15381337 Review.
-
Autonomic nerves in the mammalian choroid plexus and their influence on the formation of cerebrospinal fluid.J Cereb Blood Flow Metab. 1981;1(3):245-66. doi: 10.1038/jcbfm.1981.30. J Cereb Blood Flow Metab. 1981. PMID: 6276421 Review.
Cited by
-
The role of motile cilia in the development and physiology of the nervous system.Philos Trans R Soc Lond B Biol Sci. 2020 Feb 17;375(1792):20190156. doi: 10.1098/rstb.2019.0156. Epub 2019 Dec 30. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 31884916 Free PMC article. Review.
-
ERK signaling expands mammalian cortical radial glial cells and extends the neurogenic period.Proc Natl Acad Sci U S A. 2024 Mar 26;121(13):e2314802121. doi: 10.1073/pnas.2314802121. Epub 2024 Mar 18. Proc Natl Acad Sci U S A. 2024. PMID: 38498715 Free PMC article.
-
Evolutionary development of embryonic cerebrospinal fluid composition and regulation: an open research field with implications for brain development and function.Fluids Barriers CNS. 2016 Mar 15;13:5. doi: 10.1186/s12987-016-0029-y. Fluids Barriers CNS. 2016. PMID: 26979569 Free PMC article. Review.
-
Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2016950118. doi: 10.1073/pnas.2016950118. Proc Natl Acad Sci U S A. 2021. PMID: 33542154 Free PMC article.
-
Targeting the Choroid Plexuses for Protein Drug Delivery.Pharmaceutics. 2020 Oct 14;12(10):963. doi: 10.3390/pharmaceutics12100963. Pharmaceutics. 2020. PMID: 33066423 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
