Myocyte enhancer factor (MEF)-2 plays essential roles in T-cell transformation associated with HTLV-1 infection by stabilizing complex between Tax and CREB
- PMID: 25809782
- PMCID: PMC4374383
- DOI: 10.1186/s12977-015-0140-1
Myocyte enhancer factor (MEF)-2 plays essential roles in T-cell transformation associated with HTLV-1 infection by stabilizing complex between Tax and CREB
Abstract
Background: The exact molecular mechanisms regarding HTLV-1 Tax-mediated viral gene expression and CD4 T-cell transformation have yet to be fully delineated. Herein, utilizing virus-infected primary CD4+ T cells and the virus-producing cell line, MT-2, we describe the involvement and regulation of Myocyte enhancer factor-2 (specifically MEF-2A) during the course of HTLV-1 infection and associated disease syndrome.
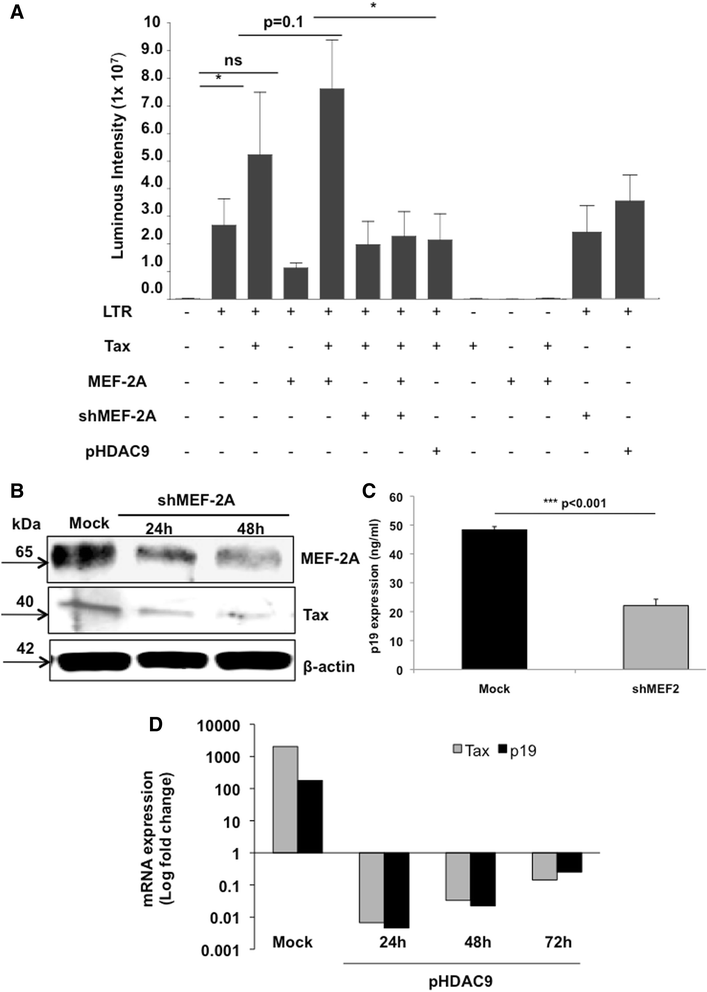
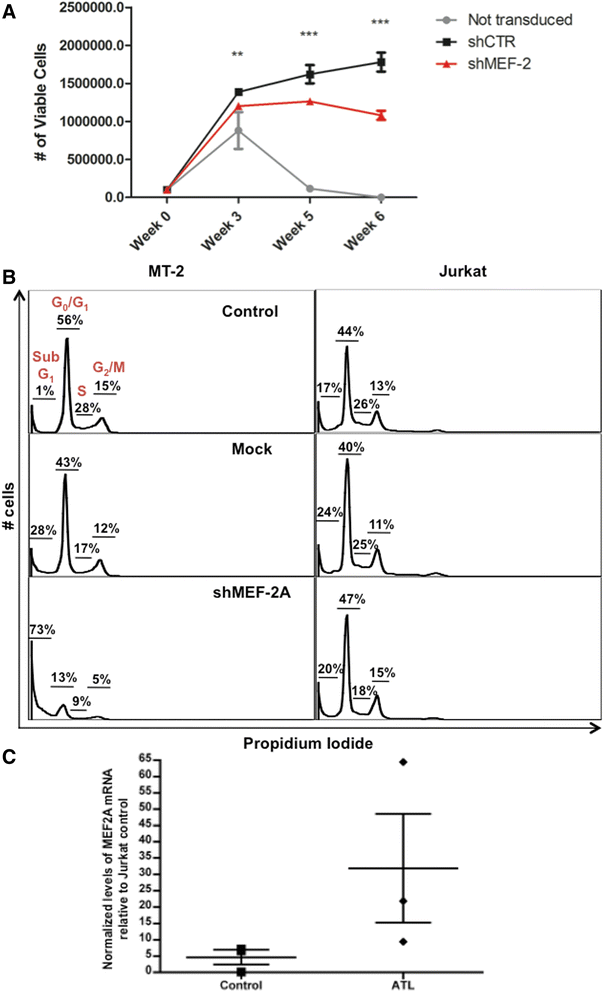
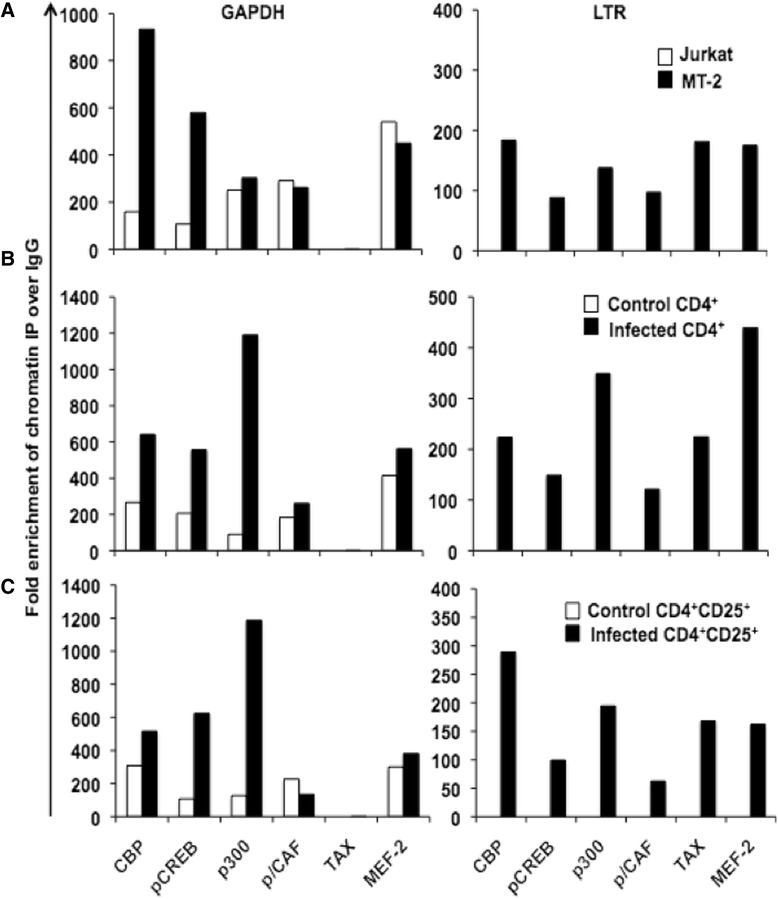
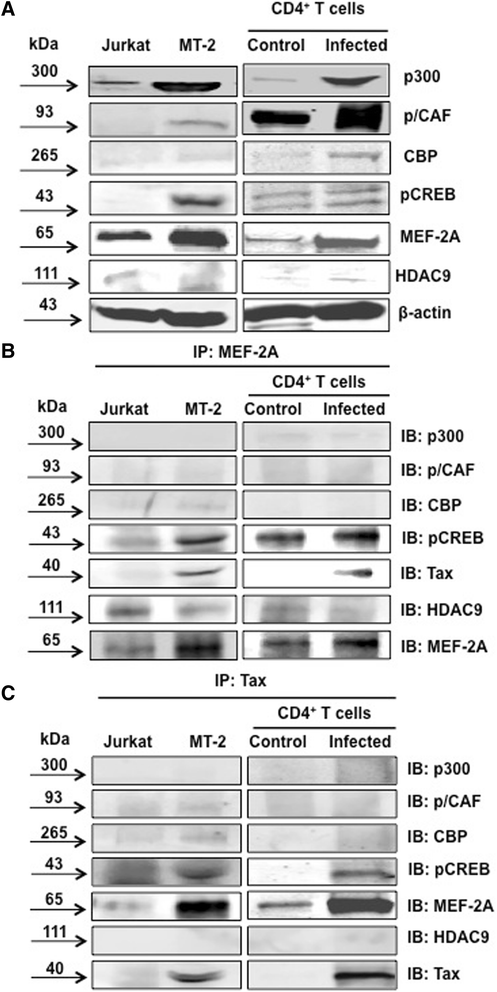
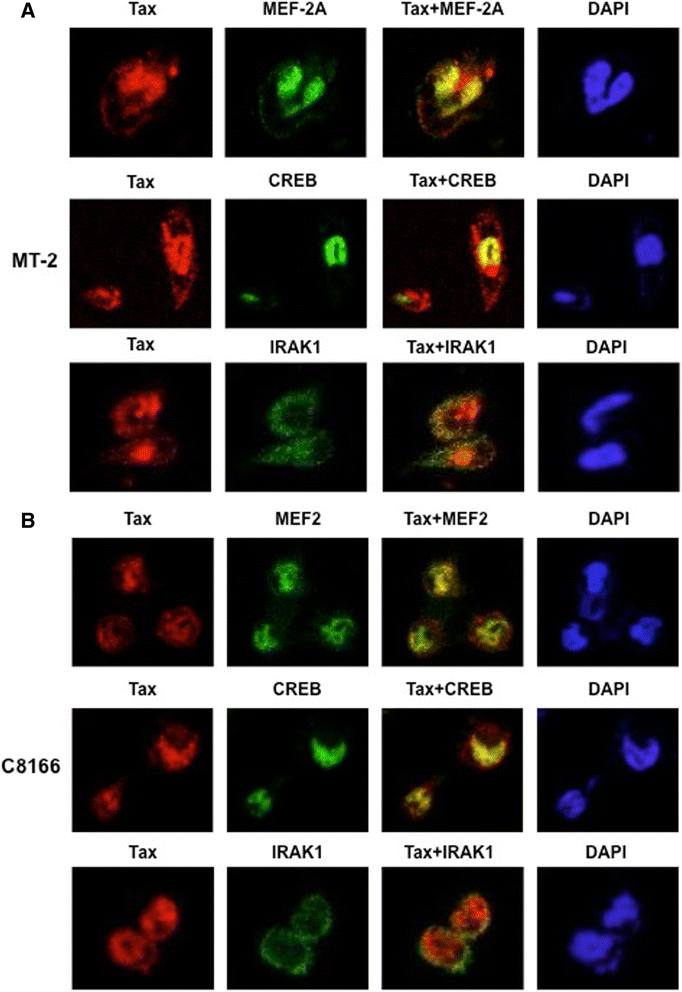
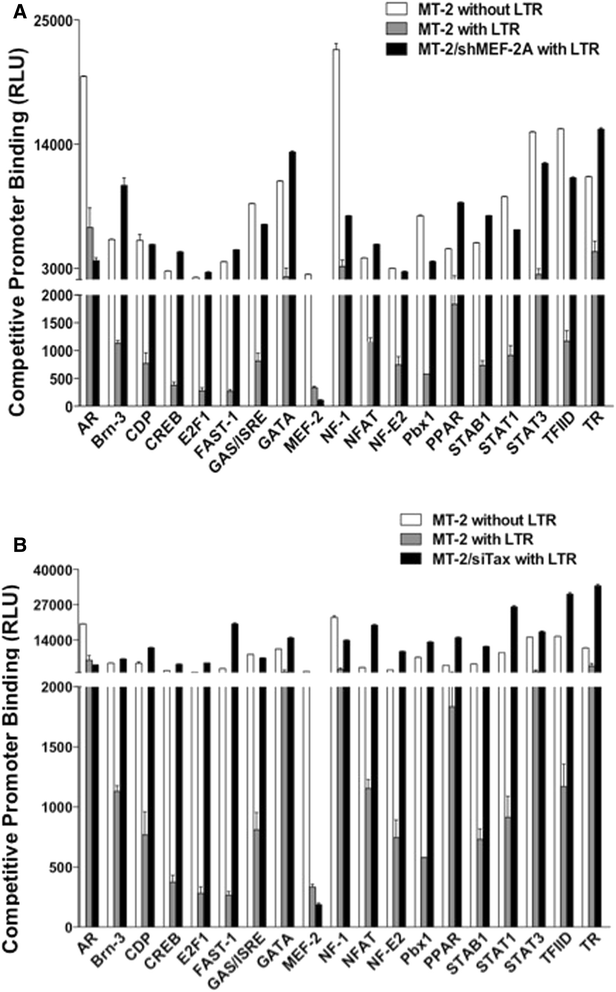
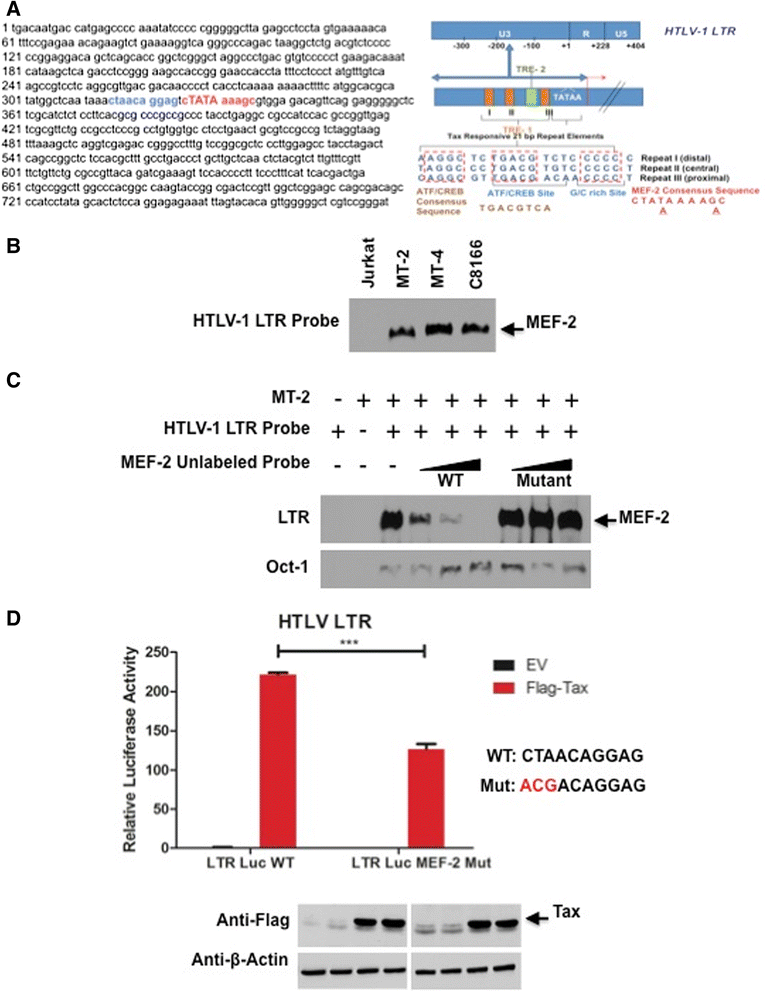
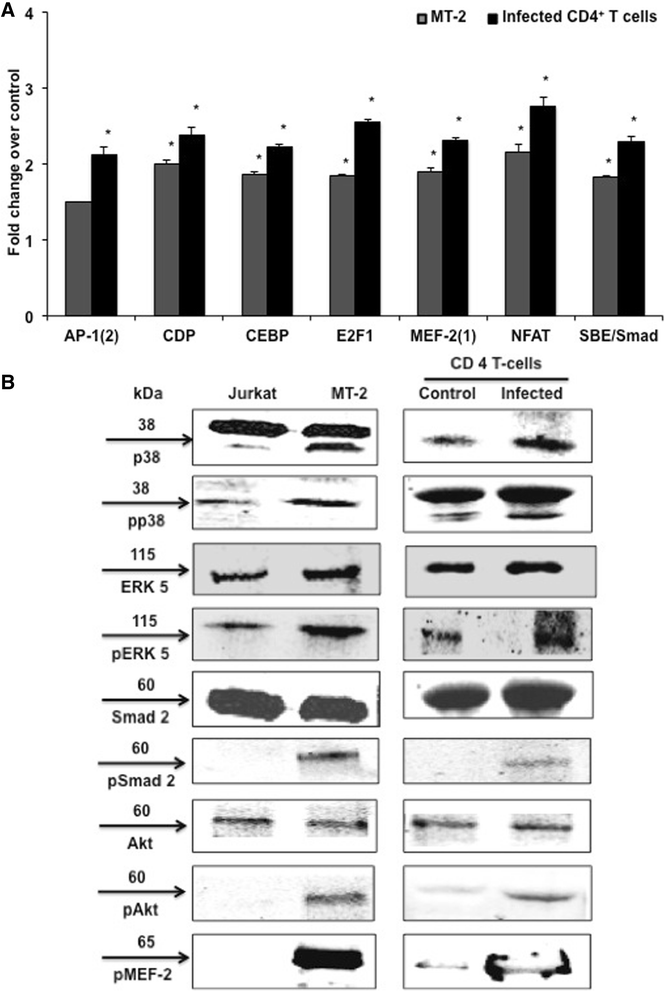
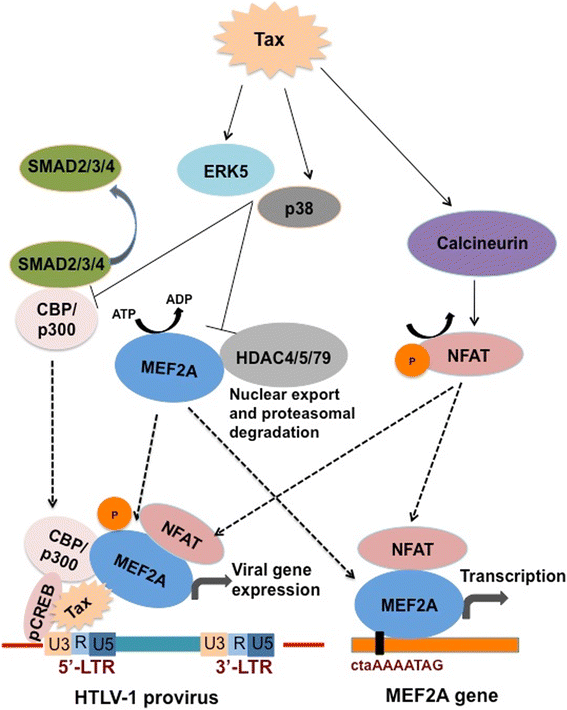
Results: Inhibition of MEF-2 expression by shRNA and its activity by HDAC9 led to reduced viral replication and T-cell transformation in correlation with a heightened expression of MEF-2 in ATL patients. Mechanistically, MEF-2 was recruited to the viral promoter (LTR, long terminal repeat) in the context of chromatin, and constituted Tax/CREB transcriptional complex via direct binding to the HTLV-1 LTR. Furthermore, an increase in MEF-2 expression was observed upon infection in an extent similar to CREB (known Tax-interacting transcription factor), and HATs (p300, CBP, and p/CAF). Confocal imaging confirmed MEF-2 co-localization with Tax and these proteins were also shown to interact by co-immunoprecipitation. MEF-2 stabilization of Tax/CREB complex was confirmed by a novel promoter-binding assay that highlighted the involvement of NFAT (nuclear factor of activated T cells) in this process via Tax-mediated activation of calcineurin (a calcium-dependent serine-threonine phosphatase). MEF-2-integrated signaling pathways (PI3K/Akt, NF-κB, MAPK, JAK/STAT, and TGF-β) were also activated during HTLV-1 infection of primary CD4+ T cells, possibly regulating MEF-2 activity.
Conclusions: We demonstrate the involvement of MEF-2 in Tax-mediated LTR activation, viral replication, and T-cell transformation in correlation with its heightened expression in ATL patients through direct binding to DNA within the HTLV-1 LTR.
Figures
Similar articles
-
Tax abolishes histone H1 repression of p300 acetyltransferase activity at the human T-cell leukemia virus type 1 promoter.J Virol. 2006 Nov;80(21):10542-53. doi: 10.1128/JVI.00631-06. Epub 2006 Aug 30. J Virol. 2006. PMID: 16943293 Free PMC article.
-
Hijacking of the O-GlcNAcZYME complex by the HTLV-1 Tax oncoprotein facilitates viral transcription.PLoS Pathog. 2017 Jul 24;13(7):e1006518. doi: 10.1371/journal.ppat.1006518. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28742148 Free PMC article.
-
Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article.
-
[Adult T-cell leukemia induced by HTLV-1: before and after HBZ].Med Sci (Paris). 2010 Apr;26(4):391-6. doi: 10.1051/medsci/2010264391. Med Sci (Paris). 2010. PMID: 20412744 Review. French.
-
Human T cell lymphotropic virus type I genomic expression and impact on intracellular signaling pathways during neurodegenerative disease and leukemia.Front Biosci. 2000 Jan 1;5:D138-68. doi: 10.2741/yao. Front Biosci. 2000. PMID: 10702386 Review.
Cited by
-
Regulation of human T-cell leukemia virus type 1 antisense promoter by myocyte enhancer factor-2C in the context of adult T-cell leukemia and lymphoma.Haematologica. 2022 Dec 1;107(12):2928-2943. doi: 10.3324/haematol.2021.279542. Haematologica. 2022. PMID: 35615924 Free PMC article.
-
Progress on Ras/MAPK Signaling Research and Targeting in Blood and Solid Cancers.Cancers (Basel). 2021 Oct 10;13(20):5059. doi: 10.3390/cancers13205059. Cancers (Basel). 2021. PMID: 34680208 Free PMC article. Review.
-
Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1.Virology. 2016 Feb;489:252-68. doi: 10.1016/j.virol.2015.12.010. Epub 2016 Jan 14. Virology. 2016. PMID: 26773386 Free PMC article.
-
Mechanisms of Oncogenesis by HTLV-1 Tax.Pathogens. 2020 Jul 7;9(7):543. doi: 10.3390/pathogens9070543. Pathogens. 2020. PMID: 32645846 Free PMC article. Review.
-
VECTOR: An Integrated Correlation Network Database for the Identification of CeRNA Axes in Uveal Melanoma.Genes (Basel). 2021 Jun 29;12(7):1004. doi: 10.3390/genes12071004. Genes (Basel). 2021. PMID: 34210067 Free PMC article.
References
-
- Palutke M, Patt DJ, Weise R, Varadachari C, Wylin RF, Bishop CR, et al. T cell leukemia–lymphoma in young adults. Am J Clin Pathol. 1977;68:429–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

