Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals
- PMID: 25802409
- PMCID: PMC4408798
- DOI: 10.1261/rna.048603.114
Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals
Abstract
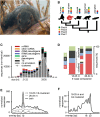
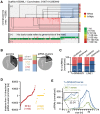
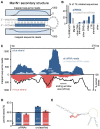
Argonaute proteins comprising Piwi-like and Argonaute-like proteins and their guiding small RNAs combat mobile DNA on the transcriptional and post-transcriptional level. While Piwi-like proteins and associated piRNAs are generally restricted to the germline, Argonaute-like proteins and siRNAs have been linked with transposon control in the germline as well as in the soma. Intriguingly, evolution has realized distinct Argonaute subfunctionalization patterns in different species but our knowledge about mammalian RNA interference pathways relies mainly on findings from the mouse model. However, mice differ from other mammals by absence of functional Piwil3 and expression of an oocyte-specific Dicer isoform. Thus, studies beyond the mouse model are required for a thorough understanding of function and evolution of mammalian RNA interference pathways. We high-throughput sequenced small RNAs from the male Tupaia belangeri germline, which represents a close outgroup to primates, hence phylogenetically links mice with humans. We identified transposon-derived piRNAs as well as siRNAs clearly contrasting the separation of piRNA- and siRNA-pathways into male and female germline as seen in mice. Genome-wide analysis of tree shrew transposons reveal that putative siRNAs map to transposon sites that form foldback secondary structures thus representing suitable Dicer substrates. In contrast piRNAs target transposon sites that remain accessible. With this we provide a basic mechanistic explanation how secondary structure of transposon transcripts influences piRNA- and siRNA-pathway utilization. Finally, our analyses of tree shrew piRNA clusters indicate A-Myb and the testis-expressed transcription factor RFX4 to be involved in the transcriptional regulation of mammalian piRNA clusters.
Keywords: RNA interference; evolution; piRNA; siRNA; transposon defense.
© 2015 Rosenkranz et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

Similar articles
-
Untangling the web: the diverse functions of the PIWI/piRNA pathway.Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
-
piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire.Genes Dev. 2015 Aug 15;29(16):1747-62. doi: 10.1101/gad.267252.115. Genes Dev. 2015. PMID: 26302790 Free PMC article.
-
A critical role for nucleoporin 358 (Nup358) in transposon silencing and piRNA biogenesis in Drosophila.J Biol Chem. 2018 Jun 15;293(24):9140-9147. doi: 10.1074/jbc.AC118.003264. Epub 2018 May 7. J Biol Chem. 2018. PMID: 29735528 Free PMC article.
-
piRNAs from Pig Testis Provide Evidence for a Conserved Role of the Piwi Pathway in Post-Transcriptional Gene Regulation in Mammals.PLoS One. 2015 May 7;10(5):e0124860. doi: 10.1371/journal.pone.0124860. eCollection 2015. PLoS One. 2015. PMID: 25950437 Free PMC article.
-
piRNA biogenesis in the germline: From transcription of piRNA genomic sources to piRNA maturation.Biochim Biophys Acta. 2016 Jan;1859(1):82-92. doi: 10.1016/j.bbagrm.2015.09.002. Epub 2015 Sep 5. Biochim Biophys Acta. 2016. PMID: 26348412 Review.
Cited by
-
Primate piRNA Cluster Evolution Suggests Limited Relevance of Pseudogenes in piRNA-Mediated Gene Regulation.Genome Biol Evol. 2019 Apr 1;11(4):1088-1104. doi: 10.1093/gbe/evz060. Genome Biol Evol. 2019. PMID: 30888404 Free PMC article.
-
Widespread selection for extremely high and low levels of secondary structure in coding sequences across all domains of life.Open Biol. 2019 May 31;9(5):190020. doi: 10.1098/rsob.190020. Epub 2019 May 29. Open Biol. 2019. PMID: 31138098 Free PMC article.
-
Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals.PLoS Genet. 2015 Nov 20;11(11):e1005652. doi: 10.1371/journal.pgen.1005652. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26588211 Free PMC article.
-
Transposable Element Targeting by piRNAs in Laurasiatherians with Distinct Transposable Element Histories.Genome Biol Evol. 2016 May 9;8(5):1327-37. doi: 10.1093/gbe/evw078. Genome Biol Evol. 2016. PMID: 27060702 Free PMC article.
-
The Research and Applications of Quantum Dots as Nano-Carriers for Targeted Drug Delivery and Cancer Therapy.Nanoscale Res Lett. 2016 Dec;11(1):207. doi: 10.1186/s11671-016-1394-9. Epub 2016 Apr 18. Nanoscale Res Lett. 2016. PMID: 27090658 Free PMC article. Review.
References
-
- Aravin AA, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al.2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
