Molecular and Functional Characterization of RecD, a Novel Member of the SF1 Family of Helicases, from Mycobacterium tuberculosis
- PMID: 25802334
- PMCID: PMC4424334
- DOI: 10.1074/jbc.M114.619395
Molecular and Functional Characterization of RecD, a Novel Member of the SF1 Family of Helicases, from Mycobacterium tuberculosis
Abstract
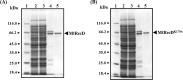
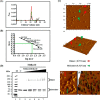
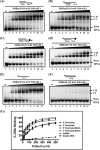
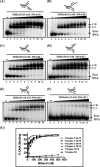
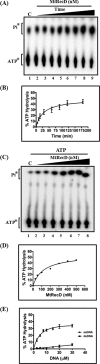
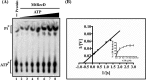
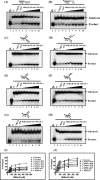
The annotated whole-genome sequence of Mycobacterium tuberculosis revealed the presence of a putative recD gene; however, the biochemical characteristics of its encoded protein product (MtRecD) remain largely unknown. Here, we show that MtRecD exists in solution as a stable homodimer. Protein-DNA binding assays revealed that MtRecD binds efficiently to single-stranded DNA and linear duplexes containing 5' overhangs relative to the 3' overhangs but not to blunt-ended duplex. Furthermore, MtRecD bound more robustly to a variety of Y-shaped DNA structures having ≥18-nucleotide overhangs but not to a similar substrate containing 5-nucleotide overhangs. MtRecD formed more salt-tolerant complexes with Y-shaped structures compared with linear duplex having 3' overhangs. The intrinsic ATPase activity of MtRecD was stimulated by single-stranded DNA. Site-specific mutagenesis of Lys-179 in motif I abolished the ATPase activity of MtRecD. Interestingly, although MtRecD-catalyzed unwinding showed a markedly higher preference for duplex substrates with 5' overhangs, it could also catalyze significant unwinding of substrates containing 3' overhangs. These results support the notion that MtRecD is a bipolar helicase with strong 5' → 3' and weak 3' → 5' unwinding activities. The extent of unwinding of Y-shaped DNA structures was ∼3-fold lower compared with duplexes with 5' overhangs. Notably, direct interaction between MtRecD and its cognate RecA led to inhibition of DNA strand exchange promoted by RecA. Altogether, these studies provide the first detailed characterization of MtRecD and present important insights into the type of DNA structure the enzyme is likely to act upon during the processes of DNA repair or homologous recombination.
Keywords: ATPase; Atomic Force Microscopy (AFM); Bioinformatics; Cloning; DNA Helicase; DNA Recombination; Enzyme Kinetics; Infectious Disease; Mycobacteria; Mycobacterium tuberculosis.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling.Biochemistry. 2009 May 19;48(19):4019-30. doi: 10.1021/bi900103d. Biochemistry. 2009. PMID: 19317511 Free PMC article.
-
Mycobacterium tuberculosis DinG is a structure-specific helicase that unwinds G4 DNA: implications for targeting G4 DNA as a novel therapeutic approach.J Biol Chem. 2014 Sep 5;289(36):25112-36. doi: 10.1074/jbc.M114.563569. Epub 2014 Jul 24. J Biol Chem. 2014. PMID: 25059658 Free PMC article.
-
Translocation by the RecB motor is an absolute requirement for {chi}-recognition and RecA protein loading by RecBCD enzyme.J Biol Chem. 2005 Nov 4;280(44):37078-87. doi: 10.1074/jbc.M505521200. Epub 2005 Jul 22. J Biol Chem. 2005. PMID: 16041060
-
A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination.FEBS Lett. 1988 Aug 1;235(1-2):16-24. doi: 10.1016/0014-5793(88)81226-2. FEBS Lett. 1988. PMID: 2841153 Free PMC article. Review.
-
Rapid purification of helicase proteins and in vitro analysis of helicase activity.Methods. 2010 Jul;51(3):322-8. doi: 10.1016/j.ymeth.2010.02.008. Epub 2010 Feb 12. Methods. 2010. PMID: 20153831 Free PMC article. Review.
Cited by
-
Prokaryotic DNA Crossroads: Holliday Junction Formation and Resolution.ACS Omega. 2024 Feb 27;9(11):12515-12538. doi: 10.1021/acsomega.3c09866. eCollection 2024 Mar 19. ACS Omega. 2024. PMID: 38524412 Free PMC article. Review.
-
Mycobacterium smegmatis HelY Is an RNA-Activated ATPase/dATPase and 3'-to-5' Helicase That Unwinds 3'-Tailed RNA Duplexes and RNA:DNA Hybrids.J Bacteriol. 2015 Oct;197(19):3057-65. doi: 10.1128/JB.00418-15. Epub 2015 Jul 13. J Bacteriol. 2015. PMID: 26170411 Free PMC article.
References
-
- Symington L. S., Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 - PubMed
-
- Pitcher R. S., Brissett N. C., Doherty A. J. (2007) Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 61, 259–282 - PubMed
-
- Cox M. M. (2007) Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42, 41–63 - PubMed
-
- Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3, D570–D603 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

