Inducible Expression of a Resistance-Nodulation-Division-Type Efflux Pump in Staphylococcus aureus Provides Resistance to Linoleic and Arachidonic Acids
- PMID: 25802299
- PMCID: PMC4420908
- DOI: 10.1128/JB.02607-14
Inducible Expression of a Resistance-Nodulation-Division-Type Efflux Pump in Staphylococcus aureus Provides Resistance to Linoleic and Arachidonic Acids
Abstract
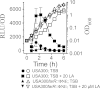
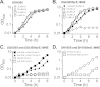
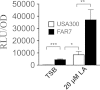
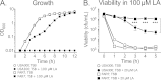
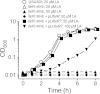
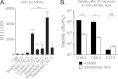
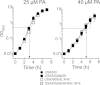
Although Staphylococcus aureus is exposed to antimicrobial fatty acids on the skin, in nasal secretions, and in abscesses, a specific mechanism of inducible resistance to this important facet of innate immunity has not been identified. Here, we have sequenced the genome of S. aureus USA300 variants selected for their ability to grow at an elevated concentration of linoleic acid. The fatty acid-resistant clone FAR7 had a single nucleotide polymorphism resulting in an H₁₂₁Y substitution in an uncharacterized transcriptional regulator belonging to the AcrR family, which was divergently transcribed from a gene encoding a member of the resistance-nodulation-division superfamily of multidrug efflux pumps. We named these genes farR and farE, for regulator and effector of fatty acid resistance, respectively. Several lines of evidence indicated that FarE promotes efflux of antimicrobial fatty acids and is regulated by FarR. First, expression of farE was strongly induced by arachidonic and linoleic acids in an farR-dependent manner. Second, an H₁₂₁Y substitution in FarR resulted in increased expression of farE and was alone sufficient to promote increased resistance of S. aureus to linoleic acid. Third, inactivation of farE resulted in a significant reduction in the inducible resistance of S. aureus to the bactericidal activity of 100 μM linoleic acid, increased accumulation of [(14)C]linoleic acid by growing cells, and severely impaired growth in the presence of nonbactericidal concentrations of linoleic acid. Cumulatively, these findings represent the first description of a specific mechanism of inducible resistance to antimicrobial fatty acids in a Gram-positive pathogen.
Importance: Staphylococcus aureus colonizes approximately 25% of humans and is a leading cause of human infectious morbidity and mortality. To persist on human hosts, S. aureus must have intrinsic defense mechanisms to cope with antimicrobial fatty acids, which comprise an important component of human innate defense mechanisms. We have identified a novel pair of genes, farR and farE, that constitute a dedicated regulator and effector of S. aureus resistance to linoleic and arachidonic acids, which are major fatty acids in human membrane phospholipid. Expression of farE, which encodes an efflux pump, is induced in an farR-dependent mechanism, in response to these antimicrobial fatty acids that would be encountered in a tissue abscess.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
DNA Binding and Sensor Specificity of FarR, a Novel TetR Family Regulator Required for Induction of the Fatty Acid Efflux Pump FarE in Staphylococcus aureus.J Bacteriol. 2019 Jan 11;201(3):e00602-18. doi: 10.1128/JB.00602-18. Print 2019 Feb 1. J Bacteriol. 2019. PMID: 30455282 Free PMC article.
-
Repeated Emergence of Variant TetR Family Regulator, FarR, and Increased Resistance to Antimicrobial Unsaturated Fatty Acid among Clonal Complex 5 Methicillin-Resistant Staphylococcus aureus.Antimicrob Agents Chemother. 2023 Mar 16;67(3):e0074922. doi: 10.1128/aac.00749-22. Epub 2023 Feb 6. Antimicrob Agents Chemother. 2023. PMID: 36744906 Free PMC article.
-
Molecular Basis of Rhodomyrtone Resistance in Staphylococcus aureus.mBio. 2021 Feb 22;13(1):e0383321. doi: 10.1128/mbio.03833-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164566 Free PMC article.
-
[The role of cell wall organization and active efflux pump systems in multidrug resistance of bacteria].Mikrobiyol Bul. 2007 Apr;41(2):309-27. Mikrobiyol Bul. 2007. PMID: 17682720 Review. Turkish.
-
Staphylococcus aureus evasion of innate antimicrobial defense.Future Microbiol. 2008 Aug;3(4):437-51. doi: 10.2217/17460913.3.4.437. Future Microbiol. 2008. PMID: 18651815 Review.
Cited by
-
Vaginal Lactobacillus fatty acid response mechanisms reveal a metabolite-targeted strategy for bacterial vaginosis treatment.Cell. 2024 Sep 19;187(19):5413-5430.e29. doi: 10.1016/j.cell.2024.07.029. Epub 2024 Aug 19. Cell. 2024. PMID: 39163861 Free PMC article.
-
The Role of Macrophages in Staphylococcus aureus Infection.Front Immunol. 2021 Jan 19;11:620339. doi: 10.3389/fimmu.2020.620339. eCollection 2020. Front Immunol. 2021. PMID: 33542723 Free PMC article. Review.
-
Arachidonic Acid Kills Staphylococcus aureus through a Lipid Peroxidation Mechanism.mBio. 2019 Oct 1;10(5):e01333-19. doi: 10.1128/mBio.01333-19. mBio. 2019. PMID: 31575763 Free PMC article.
-
The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development.Proc Natl Acad Sci U S A. 2016 Jun 21;113(25):E3538-47. doi: 10.1073/pnas.1600424113. Epub 2016 Jun 6. Proc Natl Acad Sci U S A. 2016. PMID: 27274079 Free PMC article.
-
New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria.Molecules. 2017 Mar 15;22(3):468. doi: 10.3390/molecules22030468. Molecules. 2017. PMID: 28294992 Free PMC article. Review.
References
-
- Kochanek KD, Xu J, Murphy SL, Minino AM, Kung HC. 2012. Deaths: final data for 2009. Natl Vital Stat Rep 60:1–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

