YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1
- PMID: 25800057
- PMCID: PMC4384734
- DOI: 10.1083/jcb.201411047
YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1
Abstract
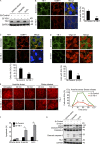
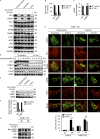
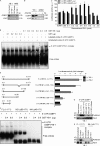
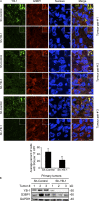
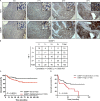
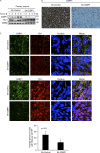
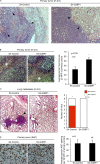
Under cell stress, global protein synthesis is inhibited to preserve energy. One mechanism is to sequester and silence mRNAs in ribonucleoprotein complexes known as stress granules (SGs), which contain translationally silent mRNAs, preinitiation factors, and RNA-binding proteins. Y-box binding protein 1 (YB-1) localizes to SGs, but its role in SG biology is unknown. We now report that YB-1 directly binds to and translationally activates the 5' untranslated region (UTR) of G3BP1 mRNAs, thereby controlling the availability of the G3BP1 SG nucleator for SG assembly. YB-1 inactivation in human sarcoma cells dramatically reduces G3BP1 and SG formation in vitro. YB-1 and G3BP1 expression are highly correlated in human sarcomas, and elevated G3BP1 expression correlates with poor survival. Finally, G3BP1 down-regulation in sarcoma xenografts prevents in vivo SG formation and tumor invasion, and completely blocks lung metastasis in mouse models. Together, these findings demonstrate a critical role for YB-1 in SG formation through translational activation of G3BP1, and highlight novel functions for SGs in tumor progression.
© 2015 Somasekharan et al.
Figures
Similar articles
-
Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression.J Virol. 2019 May 29;93(12):e00520-19. doi: 10.1128/JVI.00520-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944179 Free PMC article.
-
Translational control of aberrant stress responses as a hallmark of cancer.J Pathol. 2018 Apr;244(5):650-666. doi: 10.1002/path.5030. Epub 2018 Feb 20. J Pathol. 2018. PMID: 29293271 Review.
-
Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly.J Biol Chem. 2016 Oct 21;291(43):22671-22685. doi: 10.1074/jbc.M116.739573. Epub 2016 Sep 6. J Biol Chem. 2016. PMID: 27601476 Free PMC article.
-
G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress.Nucleic Acids Res. 2020 Jul 9;48(12):6855-6873. doi: 10.1093/nar/gkaa376. Nucleic Acids Res. 2020. PMID: 32406909 Free PMC article.
-
RNA recognition and stress granule formation by TIA proteins.Int J Mol Sci. 2014 Dec 16;15(12):23377-88. doi: 10.3390/ijms151223377. Int J Mol Sci. 2014. PMID: 25522169 Free PMC article. Review.
Cited by
-
Principles and Properties of Stress Granules.Trends Cell Biol. 2016 Sep;26(9):668-679. doi: 10.1016/j.tcb.2016.05.004. Epub 2016 Jun 9. Trends Cell Biol. 2016. PMID: 27289443 Free PMC article. Review.
-
Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer.J Hematol Oncol. 2023 Dec 18;16(1):123. doi: 10.1186/s13045-023-01522-5. J Hematol Oncol. 2023. PMID: 38110976 Free PMC article. Review.
-
Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity.Life Sci Alliance. 2018 May 16;1(2):e201800070. doi: 10.26508/lsa.201800070. eCollection 2018 May. Life Sci Alliance. 2018. PMID: 30456350 Free PMC article.
-
Phase separation drives tumor pathogenesis and evolution: all roads lead to Rome.Oncogene. 2022 Mar;41(11):1527-1535. doi: 10.1038/s41388-022-02195-z. Epub 2022 Feb 8. Oncogene. 2022. PMID: 35132182 Review.
-
Hitting the mark: Localization of mRNA and biomolecular condensates in health and disease.Wiley Interdiscip Rev RNA. 2023 Nov-Dec;14(6):e1807. doi: 10.1002/wrna.1807. Epub 2023 Jul 2. Wiley Interdiscip Rev RNA. 2023. PMID: 37393916 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

