Asymmetrically dividing Drosophila neuroblasts utilize two spatially and temporally independent cytokinesis pathways
- PMID: 25791062
- PMCID: PMC4544045
- DOI: 10.1038/ncomms7551
Asymmetrically dividing Drosophila neuroblasts utilize two spatially and temporally independent cytokinesis pathways
Abstract
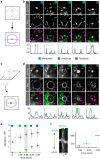
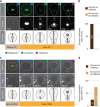
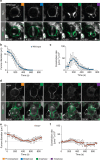
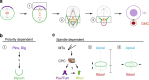
Precise cleavage furrow positioning is required for faithful chromosome segregation and cell fate determinant distribution. In most metazoan cells, contractile ring placement is regulated by the mitotic spindle through the centralspindlin complex, and potentially also the chromosomal passenger complex (CPC). Drosophila neuroblasts, asymmetrically dividing neural stem cells, but also other cells utilize both spindle-dependent and spindle-independent cleavage furrow positioning pathways. However, the relative contribution of each pathway towards cytokinesis is currently unclear. Here we report that in Drosophila neuroblasts, the mitotic spindle, but not polarity cues, controls the localization of the CPC component Survivin. We also show that Survivin and the mitotic spindle are required to stabilize the position of the cleavage furrow in late anaphase and to complete furrow constriction. These results support the model that two spatially and temporally separate pathways control different key aspects during asymmetric cell division, ensuring correct cell fate determinant segregation and neuroblast self-renewal.
Figures




Similar articles
-
Role of Survivin in cytokinesis revealed by a separation-of-function allele.Mol Biol Cell. 2011 Oct;22(20):3779-90. doi: 10.1091/mbc.E11-06-0569. Epub 2011 Aug 24. Mol Biol Cell. 2011. PMID: 21865602 Free PMC article.
-
A spindle-independent cleavage furrow positioning pathway.Nature. 2010 Sep 2;467(7311):91-4. doi: 10.1038/nature09334. Nature. 2010. PMID: 20811457 Free PMC article.
-
The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts.Development. 2001 Apr;128(7):1137-45. doi: 10.1242/dev.128.7.1137. Development. 2001. PMID: 11245579
-
Drosophila melanogaster Neuroblasts: A Model for Asymmetric Stem Cell Divisions.Results Probl Cell Differ. 2017;61:183-210. doi: 10.1007/978-3-319-53150-2_8. Results Probl Cell Differ. 2017. PMID: 28409305 Review.
-
Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy.J Cell Sci. 2005 Apr 15;118(Pt 8):1549-58. doi: 10.1242/jcs.02335. J Cell Sci. 2005. PMID: 15811947 Review.
Cited by
-
Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells.Nat Commun. 2017 Nov 9;8(1):1383. doi: 10.1038/s41467-017-01391-w. Nat Commun. 2017. PMID: 29123099 Free PMC article.
-
Phases of cortical actomyosin dynamics coupled to the neuroblast polarity cycle.Elife. 2021 Nov 15;10:e66574. doi: 10.7554/eLife.66574. Elife. 2021. PMID: 34779402 Free PMC article.
-
PAR-4 and anillin regulate myosin to coordinate spindle and furrow position during asymmetric division.J Cell Biol. 2015 Sep 28;210(7):1085-99. doi: 10.1083/jcb.201503006. J Cell Biol. 2015. PMID: 26416962 Free PMC article.
-
Actin-dependent membrane polarization reveals the mechanical nature of the neuroblast polarity cycle.Cell Rep. 2021 May 18;35(7):109146. doi: 10.1016/j.celrep.2021.109146. Cell Rep. 2021. PMID: 34010656 Free PMC article.
-
Membrane oscillations driven by Arp2/3 constrict the intercellular bridge during neural stem cell divisions.bioRxiv [Preprint]. 2024 Oct 29:2024.10.28.620743. doi: 10.1101/2024.10.28.620743. bioRxiv. 2024. PMID: 39554021 Free PMC article. Preprint.
References
-
- Lacroix B. & Maddox A. S. Cytokinesis, ploidy and aneuploidy. J. Pathol. 226, 338–351 (2012) . - PubMed
-
- Cabernard C. & Doe C. Q. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134–141 (2009) . - PubMed
-
- Cabernard C. Cytokinesis in Drosophila melanogaster. Cytoskeleton (Hoboken) 69, 791–809 (2012) . - PubMed
-
- Green R. A., Paluch E. & Oegema K. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 (2012) . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

