The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion
- PMID: 25784054
- PMCID: PMC4804868
- DOI: 10.1146/annurev-biochem-080111-092106
The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion
Abstract
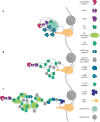
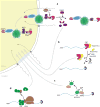
Throughout their lifetimes, messenger RNAs (mRNAs) associate with proteins to form ribonucleoproteins (mRNPs). Since the discovery of the first mRNP component more than 40 years ago, what is known as the mRNA interactome now comprises >1,000 proteins. These proteins bind mRNAs in myriad ways with varying affinities and stoichiometries, with many assembling onto nascent RNAs in a highly ordered process during transcription and precursor mRNA (pre-mRNA) processing. The nonrandom distribution of major mRNP proteins observed in transcriptome-wide studies leads us to propose that mRNPs are organized into three major domains loosely corresponding to 5' untranslated regions (UTRs), open reading frames, and 3' UTRs. Moving from the nucleus to the cytoplasm, mRNPs undergo extensive remodeling as they are first acted upon by the nuclear pore complex and then by the ribosome. When not being actively translated, cytoplasmic mRNPs can assemble into large multi-mRNP assemblies or be permanently disassembled and degraded. In this review, we aim to give the reader a thorough understanding of past and current eukaryotic mRNP research.
Keywords: RNA granules; RNA processing; RNP remodeling; coupling; mRNP packaging; ribonucleoproteins.
Figures


Similar articles
-
Integration of mRNP formation and export.Cell Mol Life Sci. 2017 Aug;74(16):2875-2897. doi: 10.1007/s00018-017-2503-3. Epub 2017 Mar 17. Cell Mol Life Sci. 2017. PMID: 28314893 Free PMC article. Review.
-
Principles and properties of eukaryotic mRNPs.Mol Cell. 2014 May 22;54(4):547-58. doi: 10.1016/j.molcel.2014.04.033. Mol Cell. 2014. PMID: 24856220 Review.
-
The interplay of nuclear mRNP assembly, mRNA surveillance and export.Trends Cell Biol. 2003 Jun;13(6):319-27. doi: 10.1016/s0962-8924(03)00106-5. Trends Cell Biol. 2003. PMID: 12791298 Review.
-
Cytoplasmic mRNA-protein complexes of chicken muscle cells and their role in protein synthesis.Eur J Biochem. 1984 Jun 1;141(2):247-54. doi: 10.1111/j.1432-1033.1984.tb08184.x. Eur J Biochem. 1984. PMID: 6734598
-
The Chironomus tentans translation initiation factor eIF4H is present in the nucleus but does not bind to mRNA until the mRNA reaches the cytoplasmic perinuclear region.J Cell Sci. 2003 Nov 15;116(Pt 22):4521-32. doi: 10.1242/jcs.00766. J Cell Sci. 2003. PMID: 14576346
Cited by
-
Functional Integration of mRNA Translational Control Programs.Biomolecules. 2015 Jul 21;5(3):1580-99. doi: 10.3390/biom5031580. Biomolecules. 2015. PMID: 26197342 Free PMC article. Review.
-
Genome-Wide Analysis of Differentially Expressed miRNAs and Their Associated Regulatory Networks in Lenses Deficient for the Congenital Cataract-Linked Tudor Domain Containing Protein TDRD7.Front Cell Dev Biol. 2021 Feb 16;9:615761. doi: 10.3389/fcell.2021.615761. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33665188 Free PMC article.
-
Dynamic mRNP Remodeling in Response to Internal and External Stimuli.Biomolecules. 2020 Sep 11;10(9):1310. doi: 10.3390/biom10091310. Biomolecules. 2020. PMID: 32932892 Free PMC article. Review.
-
Precise gene models using long-read sequencing reveal a unique poly(A) signal in Giardia lamblia.RNA. 2022 May;28(5):668-682. doi: 10.1261/rna.078793.121. Epub 2022 Feb 2. RNA. 2022. PMID: 35110372 Free PMC article.
-
Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription.Elife. 2018 Oct 3;7:e37663. doi: 10.7554/eLife.37663. Elife. 2018. PMID: 30281021 Free PMC article.
References
-
- Gall JG. On the submicroscopic structure of chromosomes. Brookhaven Symp Biol. 1956;8:17–32. - PubMed
-
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–98. - PubMed
-
- Kumar A, Pederson T. Comparison of proteins bound to heterogeneous nuclear RNA and messenger RNA in HeLa cells. J Mol Biol. 1975;96:353–65. - PubMed
-
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–38. - PubMed
-
- Barrieux A, Ingraham HA, David DN, Rosenfeld MG. Isolation of messenger-like ribonucleoproteins. Biochemistry. 1975;14:1815–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

