Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers
- PMID: 25779043
- PMCID: PMC4402534
- DOI: 10.1093/nar/gkv215
Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers
Abstract
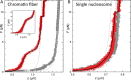
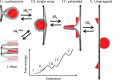
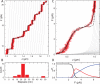
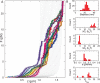
Single-molecule techniques allow for picoNewton manipulation and nanometer accuracy measurements of single chromatin fibers. However, the complexity of the data, the heterogeneity of the composition of individual fibers and the relatively large fluctuations in extension of the fibers complicate a structural interpretation of such force-extension curves. Here we introduce a statistical mechanics model that quantitatively describes the extension of individual fibers in response to force on a per nucleosome basis. Four nucleosome conformations can be distinguished when pulling a chromatin fiber apart. A novel, transient conformation is introduced that coexists with single wrapped nucleosomes between 3 and 7 pN. Comparison of force-extension curves between single nucleosomes and chromatin fibers shows that embedding nucleosomes in a fiber stabilizes the nucleosome by 10 kBT. Chromatin fibers with 20- and 50-bp linker DNA follow a different unfolding pathway. These results have implications for accessibility of DNA in fully folded and partially unwrapped chromatin fibers and are vital for understanding force unfolding experiments on nucleosome arrays.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
10 years of tension on chromatin: results from single molecule force spectroscopy.Curr Pharm Biotechnol. 2009 Aug;10(5):474-85. doi: 10.2174/138920109788922128. Curr Pharm Biotechnol. 2009. PMID: 19689315 Review.
-
Characterization of nucleosome unwrapping within chromatin fibers using magnetic tweezers.Biophys J. 2014 Jul 15;107(2):373-383. doi: 10.1016/j.bpj.2014.05.036. Biophys J. 2014. PMID: 25028879 Free PMC article.
-
Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber.Nat Struct Mol Biol. 2009 May;16(5):534-40. doi: 10.1038/nsmb.1590. Epub 2009 Apr 19. Nat Struct Mol Biol. 2009. PMID: 19377481
-
Rigid Basepair Monte Carlo Simulations of One-Start and Two-Start Chromatin Fiber Unfolding by Force.Biophys J. 2018 Nov 20;115(10):1848-1859. doi: 10.1016/j.bpj.2018.10.007. Epub 2018 Oct 11. Biophys J. 2018. PMID: 30366627 Free PMC article.
-
Chromatin under mechanical stress: from single 30 nm fibers to single nucleosomes.FEBS J. 2011 Jul;278(13):2231-43. doi: 10.1111/j.1742-4658.2011.08153.x. Epub 2011 May 26. FEBS J. 2011. PMID: 21535477 Review.
Cited by
-
Nucleosome plasticity is a critical element of chromatin liquid-liquid phase separation and multivalent nucleosome interactions.Nat Commun. 2021 May 17;12(1):2883. doi: 10.1038/s41467-021-23090-3. Nat Commun. 2021. PMID: 34001913 Free PMC article.
-
DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability.Nat Commun. 2020 Jun 5;11(1):2828. doi: 10.1038/s41467-020-16702-x. Nat Commun. 2020. PMID: 32504003 Free PMC article.
-
Torsional stress can regulate the unwrapping of two outer half superhelical turns of nucleosomal DNA.Proc Natl Acad Sci U S A. 2021 Feb 16;118(7):e2020452118. doi: 10.1073/pnas.2020452118. Proc Natl Acad Sci U S A. 2021. PMID: 33558240 Free PMC article.
-
Bottom-Up Meets Top-Down: The Crossroads of Multiscale Chromatin Modeling.Biophys J. 2020 May 5;118(9):2057-2065. doi: 10.1016/j.bpj.2020.03.014. Epub 2020 Apr 4. Biophys J. 2020. PMID: 32320675 Free PMC article. Review.
-
Chromatin fiber breaks into clutches under tension and crowding.Nucleic Acids Res. 2022 Sep 23;50(17):9738-9747. doi: 10.1093/nar/gkac725. Nucleic Acids Res. 2022. PMID: 36029149 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

