IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes
- PMID: 25775030
- PMCID: PMC4363081
- DOI: 10.1038/ncomms7379
IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes
Abstract
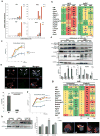
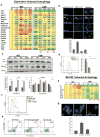
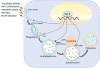
Autophagy, activated by many stresses, plays a critical role in innate immune responses. Here we show that interferon regulatory factor 8 (IRF8) is required for the expression of autophagy-related genes in dendritic cells. Furthermore in macrophages, IRF8 is induced by multiple autophagy-inducing stresses, including IFNγ and Toll-like receptor stimulation, bacterial infection, starvation and by macrophage colony-stimulating factor. IRF8 directly activates many genes involved in various steps of autophagy, promoting autophagosome formation and lysosomal fusion. Consequently, Irf8(-/-) macrophages are deficient in autophagic activity, and excessively accumulate SQSTM1 and ubiquitin-bound proteins. We show that clearance of Listeria monocytogenes in macrophages requires IRF8-dependent activation of autophagy genes and subsequent autophagic capturing and degradation of Listeria antigens. These processes are defective in Irf8(-/-) macrophages where uninhibited bacterial growth ensues. Together these data suggest that IRF8 is a major autophagy regulator in macrophages, essential for macrophage maturation, survival and innate immune responses.
Conflict of interest statement
Figures

Similar articles
-
Interferon regulatory factor 8 regulates pathways for antigen presentation in myeloid cells and during tuberculosis.PLoS Genet. 2011 Jun;7(6):e1002097. doi: 10.1371/journal.pgen.1002097. Epub 2011 Jun 23. PLoS Genet. 2011. PMID: 21731497 Free PMC article.
-
Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation.Blood. 2013 Mar 7;121(10):1839-49. doi: 10.1182/blood-2012-06-437863. Epub 2013 Jan 14. Blood. 2013. PMID: 23319570 Free PMC article.
-
PU.1 and ICSBP control constitutive and IFN-gamma-regulated Tlr9 gene expression in mouse macrophages.J Leukoc Biol. 2007 Jun;81(6):1577-90. doi: 10.1189/jlb.0107036. Epub 2007 Mar 14. J Leukoc Biol. 2007. PMID: 17360957
-
Genetic determinants of susceptibility to Mycobacterial infections: IRF8, a new kid on the block.Adv Exp Med Biol. 2013;783:45-80. doi: 10.1007/978-1-4614-6111-1_3. Adv Exp Med Biol. 2013. PMID: 23468103 Review.
-
Interferon regulatory factor 8 governs myeloid cell development.Cytokine Growth Factor Rev. 2020 Oct;55:48-57. doi: 10.1016/j.cytogfr.2020.03.003. Epub 2020 Apr 12. Cytokine Growth Factor Rev. 2020. PMID: 32327344 Review.
Cited by
-
Murine IRF8 Mutation Offers New Insight into Osteoclast and Root Resorption.J Dent Res. 2024 Mar;103(3):318-328. doi: 10.1177/00220345231222173. Epub 2024 Feb 12. J Dent Res. 2024. PMID: 38343385
-
Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model.Autophagy. 2019 May;15(5):796-812. doi: 10.1080/15548627.2019.1569297. Epub 2019 Jan 24. Autophagy. 2019. PMID: 30676840 Free PMC article.
-
Emerging mechanistic insights of selective autophagy in hepatic diseases.Front Pharmacol. 2023 Mar 16;14:1149809. doi: 10.3389/fphar.2023.1149809. eCollection 2023. Front Pharmacol. 2023. PMID: 37007026 Free PMC article. Review.
-
Interactions with Commensal and Pathogenic Bacteria Induce HIV-1 Latency in Macrophages through Altered Transcription Factor Recruitment to the LTR.J Virol. 2021 Mar 10;95(7):e02141-20. doi: 10.1128/JVI.02141-20. Epub 2021 Jan 20. J Virol. 2021. PMID: 33472928 Free PMC article.
-
Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages.Front Immunol. 2015 May 26;6:263. doi: 10.3389/fimmu.2015.00263. eCollection 2015. Front Immunol. 2015. PMID: 26074923 Free PMC article. Review.
References
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

