Whole-genome analysis revealed the positively selected genes during the differentiation of indica and temperate japonica rice
- PMID: 25774680
- PMCID: PMC4361536
- DOI: 10.1371/journal.pone.0119239
Whole-genome analysis revealed the positively selected genes during the differentiation of indica and temperate japonica rice
Abstract
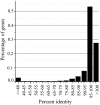
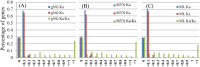
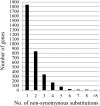
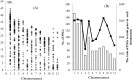
To investigate the selective pressures acting on the protein-coding genes during the differentiation of indica and japonica, all of the possible orthologous genes between the Nipponbare and 93-11 genomes were identified and compared with each other. Among these genes, 8,530 pairs had identical sequences, and 27,384 pairs shared more than 90% sequence identity. Only 2,678 pairs of genes displaying a Ka/Ks ratio significantly greater than one were revealed, and most of these genes contained only nonsynonymous sites. The genes without synonymous site were further analyzed with the SNP data of 1529 O. sativa and O. rufipogon accessions, and 1068 genes were identified to be under positive selection during the differentiation of indica and temperate japonica. The positively selected genes (PSGs) are unevenly distributed on 12 chromosomes, and the proteins encoded by the PSGs are dominant with binding, transferase and hydrolase activities, and especially enriched in the plant responses to stimuli, biological regulations, and transport processes. Meanwhile, the most PSGs of the known function and/or expression were involved in the regulation of biotic/abiotic stresses. The evidence of pervasive positive selection suggested that many factors drove the differentiation of indica and japonica, which has already started in wild rice but is much lower than in cultivated rice. Lower differentiation and less PSGs revealed between the Or-It and Or-IIIt wild rice groups implied that artificial selection provides greater contribution on the differentiation than natural selection. In addition, the phylogenetic tree constructed with positively selected sites showed that the japonica varieties exhibited more diversity than indica on differentiation, and Or-III of O. rufipogon exhibited more than Or-I.
Conflict of interest statement
Figures
Similar articles
-
Chloroplast DNA polymorphism and evolutional relationships between Asian cultivated rice (Oryza sativa) and its wild relatives (O. rufipogon).Genet Mol Res. 2012 Dec 17;11(4):4418-31. doi: 10.4238/2012.October.9.8. Genet Mol Res. 2012. PMID: 23096910
-
Molecular evolution of the TAC1 gene from rice (Oryza sativa L.).J Genet Genomics. 2012 Oct 20;39(10):551-60. doi: 10.1016/j.jgg.2012.07.011. Epub 2012 Sep 16. J Genet Genomics. 2012. PMID: 23089365
-
Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice.Theor Appl Genet. 2007 Feb;114(4):731-43. doi: 10.1007/s00122-006-0473-1. Epub 2007 Jan 12. Theor Appl Genet. 2007. PMID: 17219210
-
Natural variation of rice blast resistance gene Pi-d2.Genet Mol Res. 2015 Feb 13;14(1):1235-49. doi: 10.4238/2015.February.13.2. Genet Mol Res. 2015. PMID: 25730062
-
Rice ( Oryza) hemoglobins.F1000Res. 2014 Oct 27;3:253. doi: 10.12688/f1000research.5530.2. eCollection 2014. F1000Res. 2014. PMID: 25653837 Free PMC article. Review.
Cited by
-
The ties of brotherhood between japonica and indica rice for regional adaptation.Sci China Life Sci. 2022 Jul;65(7):1369-1379. doi: 10.1007/s11427-021-2019-x. Epub 2021 Dec 9. Sci China Life Sci. 2022. PMID: 34902099
-
Genome-Wide Association Mapping to Identify Genetic Loci for Cold Tolerance and Cold Recovery During Germination in Rice.Front Genet. 2020 Feb 21;11:22. doi: 10.3389/fgene.2020.00022. eCollection 2020. Front Genet. 2020. PMID: 32153631 Free PMC article.
-
Selective sweep with significant positive selection serves as the driving force for the differentiation of japonica and indica rice cultivars.BMC Genomics. 2017 Apr 19;18(1):307. doi: 10.1186/s12864-017-3702-x. BMC Genomics. 2017. PMID: 28420345 Free PMC article.
-
Candidate loci involved in domestication and improvement detected by a published 90K wheat SNP array.Sci Rep. 2017 Mar 22;7:44530. doi: 10.1038/srep44530. Sci Rep. 2017. PMID: 28327671 Free PMC article.
-
High-Quality Genomes and High-Density Genetic Map Facilitate the Identification of Genes From a Weedy Rice.Front Plant Sci. 2021 Nov 19;12:775051. doi: 10.3389/fpls.2021.775051. eCollection 2021. Front Plant Sci. 2021. PMID: 34868173 Free PMC article.
References
-
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant molecular biology. 1997;35(1–2):25–34. Epub 1997/09/18 - PubMed
-
- Oka HI, editor. Origin of cultivated rice. Tokyo: Japan Scientific Societies Press; 1988.
-
- Morishima H, Oka. H . Phylogenetic differentiation of cultivated rice. XXII. Numerical evaluation of the Indica—Japonica differentiation. Jpn J Breed. 1981;31:402–13.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

