A Powerful New Quantitative Genetics Platform, Combining Caenorhabditis elegans High-Throughput Fitness Assays with a Large Collection of Recombinant Strains
- PMID: 25770127
- PMCID: PMC4426375
- DOI: 10.1534/g3.115.017178
A Powerful New Quantitative Genetics Platform, Combining Caenorhabditis elegans High-Throughput Fitness Assays with a Large Collection of Recombinant Strains
Abstract
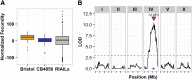
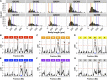
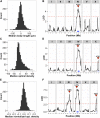
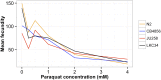
The genetic variants underlying complex traits are often elusive even in powerful model organisms such as Caenorhabditis elegans with controlled genetic backgrounds and environmental conditions. Two major contributing factors are: (1) the lack of statistical power from measuring the phenotypes of small numbers of individuals, and (2) the use of phenotyping platforms that do not scale to hundreds of individuals and are prone to noisy measurements. Here, we generated a new resource of 359 recombinant inbred strains that augments the existing C. elegans N2xCB4856 recombinant inbred advanced intercross line population. This new strain collection removes variation in the neuropeptide receptor gene npr-1, known to have large physiological and behavioral effects on C. elegans and mitigates the hybrid strain incompatibility caused by zeel-1 and peel-1, allowing for identification of quantitative trait loci that otherwise would have been masked by those effects. Additionally, we optimized highly scalable and accurate high-throughput assays of fecundity and body size using the COPAS BIOSORT large particle nematode sorter. Using these assays, we identified quantitative trait loci involved in fecundity and growth under normal growth conditions and after exposure to the herbicide paraquat, including independent genetic loci that regulate different stages of larval growth. Our results offer a powerful platform for the discovery of the genetic variants that control differences in responses to drugs, other aqueous compounds, bacterial foods, and pathogenic stresses.
Keywords: C. elegans; QTL mapping; fitness assays; high-throughput phenotyping.
Copyright © 2015 Andersen et al.
Figures

Similar articles
-
Linkage mapping reveals loci that underlie differences in Caenorhabditis elegans growth.G3 (Bethesda). 2022 Sep 30;12(10):jkac207. doi: 10.1093/g3journal/jkac207. G3 (Bethesda). 2022. PMID: 35961034 Free PMC article.
-
Shared Genomic Regions Underlie Natural Variation in Diverse Toxin Responses.Genetics. 2018 Dec;210(4):1509-1525. doi: 10.1534/genetics.118.301311. Epub 2018 Oct 19. Genetics. 2018. PMID: 30341085 Free PMC article.
-
Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1.BMC Genomics. 2016 Apr 11;17:280. doi: 10.1186/s12864-016-2603-8. BMC Genomics. 2016. PMID: 27066825 Free PMC article.
-
Caenorhabditis elegans as a platform for molecular quantitative genetics and the systems biology of natural variation.Genet Res (Camb). 2010 Dec;92(5-6):331-48. doi: 10.1017/S0016672310000601. Genet Res (Camb). 2010. PMID: 21429266 Review.
-
Natural genetic variation as a tool for discovery in Caenorhabditis nematodes.Genetics. 2022 Jan 4;220(1):iyab156. doi: 10.1093/genetics/iyab156. Genetics. 2022. PMID: 35134197 Free PMC article. Review.
Cited by
-
The hypoxia-response pathway modulates RAS/MAPK-mediated cell fate decisions in Caenorhabditis elegans.Life Sci Alliance. 2019 May 24;2(3):e201800255. doi: 10.26508/lsa.201800255. Print 2019 Jun. Life Sci Alliance. 2019. PMID: 31126994 Free PMC article.
-
Linkage mapping reveals loci that underlie differences in Caenorhabditis elegans growth.G3 (Bethesda). 2022 Sep 30;12(10):jkac207. doi: 10.1093/g3journal/jkac207. G3 (Bethesda). 2022. PMID: 35961034 Free PMC article.
-
Experimental Evolution with Caenorhabditis Nematodes.Genetics. 2017 Jun;206(2):691-716. doi: 10.1534/genetics.115.186288. Genetics. 2017. PMID: 28592504 Free PMC article. Review.
-
Hierarchical compression of Caenorhabditis elegans locomotion reveals phenotypic differences in the organization of behaviour.J R Soc Interface. 2016 Aug;13(121):20160466. doi: 10.1098/rsif.2016.0466. J R Soc Interface. 2016. PMID: 27581484 Free PMC article.
-
Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism.Elife. 2019 Apr 8;8:e40260. doi: 10.7554/eLife.40260. Elife. 2019. PMID: 30958264 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
