DNA methylation and childhood asthma in the inner city
- PMID: 25769910
- PMCID: PMC4494877
- DOI: 10.1016/j.jaci.2015.01.025
DNA methylation and childhood asthma in the inner city
Abstract
Background: Epigenetic marks are heritable, influenced by the environment, direct the maturation of T lymphocytes, and in mice enhance the development of allergic airway disease. Thus it is important to define epigenetic alterations in asthmatic populations.
Objective: We hypothesize that epigenetic alterations in circulating PBMCs are associated with allergic asthma.
Methods: We compared DNA methylation patterns and gene expression in inner-city children with persistent atopic asthma versus healthy control subjects by using DNA and RNA from PBMCs. Results were validated in an independent population of asthmatic patients.
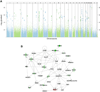
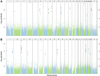
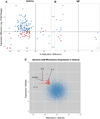
Results: Comparing asthmatic patients (n = 97) with control subjects (n = 97), we identified 81 regions that were differentially methylated. Several immune genes were hypomethylated in asthma, including IL13, RUNX3, and specific genes relevant to T lymphocytes (TIGIT). Among asthmatic patients, 11 differentially methylated regions were associated with higher serum IgE concentrations, and 16 were associated with percent predicted FEV1. Hypomethylated and hypermethylated regions were associated with increased and decreased gene expression, respectively (P < 6 × 10(-12) for asthma and P < .01 for IgE). We further explored the relationship between DNA methylation and gene expression using an integrative analysis and identified additional candidates relevant to asthma (IL4 and ST2). Methylation marks involved in T-cell maturation (RUNX3), TH2 immunity (IL4), and oxidative stress (catalase) were validated in an independent asthmatic cohort of children living in the inner city.
Conclusions: Our results demonstrate that DNA methylation marks in specific gene loci are associated with asthma and suggest that epigenetic changes might play a role in establishing the immune phenotype associated with asthma.
Keywords: DNA methylation; T(H)2 immunity; atopic asthma; epigenetics; inner city.
Published by Elsevier Inc.
Conflict of interest statement
Disclosure of potential conflict of interest: I. V. Yang has received research support from the National Institutes of Health (NIH). A. Liu has received payment for lectures from Merck and is on the data safety monitoring committee from GlaxoSmithKline. G. T. O’Connor has received research support from the NIH. S. J. Teach has received research and travel support from the NIH/National Institute of Allergy and Infectious Diseases (NIAID), Patient-Centered Outcome Research Institute, Fight for Children, the DC Department of Health, and the Kellogg Foundation; is employed by Children’s National Health System; and receives royalties from Up-To-Date. M. Kattan has received research support from the NIH and is a member of the Novartis Advisory Board. R. Gruchalla has received research and travel support from the NIAID. S. J. Szefler has received research support from the NIAID and GlaxoSmithKline; has consultant arrangements with Merck, Boehringer Ingelheim, GlaxoSmithKline, and Genentech; has received payment for lectures from Merck; and has submitted a patent for β-adrenengic receptor polymorphism for the National Heart, Lung, and Blood Institute CARE Network. M. A. Gill has received research support from the NIH/ NIAID. A. Calatroni has received research support from the NIH/NIAID. G. David has a contract with the NIH/NIAID. W. W. Busse has received research support from the NIH/NIAID and the National Heart, Lung, and Blood Institute; is a board member for Merck; has consultant arrangements with Novartis, GlaxoSmithKline, Roche, Pfizer, Boston Scientific, Circassia, ICON, AstraZeneca, Sanofi, Amgen, Med-Immune, NeoStem, Takeda, and Boehringer Ingelheim; and has received royalties from Elsevier. D. A. Schwartz has received research support from the NIH and the Veterans Administration; has consultant arrangements with Novartis and Boehringer-Ingelheim; is employed by the University of Colorado Medical School and the Department of Veterans Affairs; has provided expert testimony from Weitz and Luxenberg Law Firm, Brayton and Purcell Law Firm, and Wallace and Graham Law Firm; has patents for TLR2 single nucleotide polymorphism, MUC5b single nucleotide polymorphism, and has patent applications (61/248,505, 61/666,233, 60/ 992,079); and has received royalties from Springer. The rest of the authors declare that they have no relevant conflicts of interest.
Figures
Similar articles
-
The nasal methylome and childhood atopic asthma.J Allergy Clin Immunol. 2017 May;139(5):1478-1488. doi: 10.1016/j.jaci.2016.07.036. Epub 2016 Oct 13. J Allergy Clin Immunol. 2017. PMID: 27745942 Free PMC article.
-
DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children.PLoS One. 2012;7(9):e44213. doi: 10.1371/journal.pone.0044213. Epub 2012 Sep 6. PLoS One. 2012. PMID: 22970180 Free PMC article.
-
Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy.Allergy. 2013 Mar;68(3):355-64. doi: 10.1111/all.12097. Epub 2013 Jan 25. Allergy. 2013. PMID: 23346934
-
Epigenetic mechanisms and the development of asthma.J Allergy Clin Immunol. 2012 Dec;130(6):1243-55. doi: 10.1016/j.jaci.2012.07.052. Epub 2012 Sep 29. J Allergy Clin Immunol. 2012. PMID: 23026498 Free PMC article. Review.
-
Cell-Specific DNA Methylation Signatures in Asthma.Genes (Basel). 2019 Nov 15;10(11):932. doi: 10.3390/genes10110932. Genes (Basel). 2019. PMID: 31731604 Free PMC article. Review.
Cited by
-
Epigenome-wide association study on asthma and chronic obstructive pulmonary disease overlap reveals aberrant DNA methylations related to clinical phenotypes.Sci Rep. 2021 Mar 3;11(1):5022. doi: 10.1038/s41598-021-83185-1. Sci Rep. 2021. PMID: 33658578 Free PMC article.
-
DNA methylation and smoking in Korean adults: epigenome-wide association study.Clin Epigenetics. 2016 Sep 22;8:103. doi: 10.1186/s13148-016-0266-6. eCollection 2016. Clin Epigenetics. 2016. PMID: 27688819 Free PMC article.
-
DNA methylation biomarkers in asthma and rhinitis: Are we there yet?Clin Transl Allergy. 2022 Mar;12(3):e12131. doi: 10.1002/clt2.12131. Clin Transl Allergy. 2022. PMID: 35344303 Free PMC article. Review.
-
Inner-City Asthma in Children.Clin Rev Allergy Immunol. 2019 Apr;56(2):248-268. doi: 10.1007/s12016-019-08728-x. Clin Rev Allergy Immunol. 2019. PMID: 30666508 Review.
-
The nasal methylome as a biomarker of asthma and airway inflammation in children.Nat Commun. 2019 Jul 12;10(1):3095. doi: 10.1038/s41467-019-11058-3. Nat Commun. 2019. PMID: 31300640 Free PMC article.
References
-
- Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. - PubMed
-
- Lockett GA, Holloway JW. Genome-wide association studies in asthma; perhaps, the end of the beginning. Curr Opin Allergy Clin Immunol. 2013;13:463–469. - PubMed
-
- Weiss ST, Silverman EK. Pro: genome-wide association studies (GWAS) in asthma. Am J Respir Crit Care Med. 2011;184:631–633. - PubMed
-
- Moffatt MF, Cookson WO. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy. 1998;28(suppl 1):56–61. discussion 5–6. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

