Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution
- PMID: 25765714
- PMCID: PMC4333152
- DOI: 10.1186/s12864-015-1245-6
Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution
Abstract
Background: Because species-specific gene expression is driven by species-specific regulation, understanding the relationship between sequence and function of the regulatory regions in different species will help elucidate how differences among species arise. Despite active experimental and computational research, relationships among sequence, conservation, and function are still poorly understood.
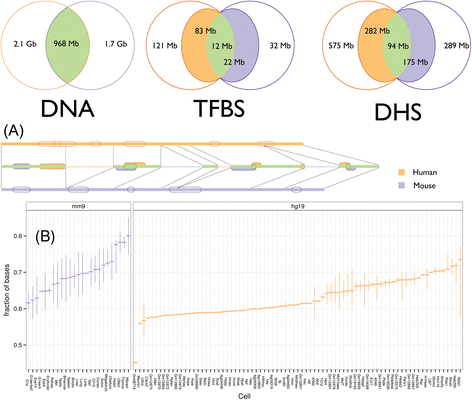
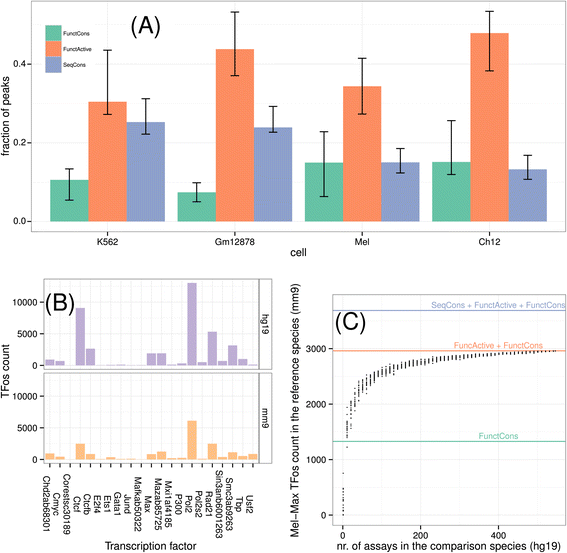
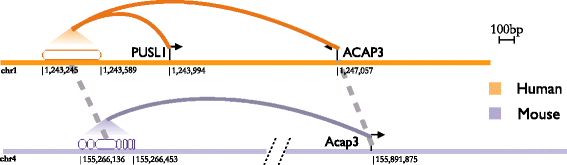
Results: We compared transcription factor occupied segments (TFos) for 116 human and 35 mouse TFs in 546 human and 125 mouse cell types and tissues from the Human and the Mouse ENCODE projects. We based the map between human and mouse TFos on a one-to-one nucleotide cross-species mapper, bnMapper, that utilizes whole genome alignments (WGA). Our analysis shows that TFos are under evolutionary constraint, but a substantial portion (25.1% of mouse and 25.85% of human on average) of the TFos does not have a homologous sequence on the other species; this portion varies among cell types and TFs. Furthermore, 47.67% and 57.01% of the homologous TFos sequence shows binding activity on the other species for human and mouse respectively. However, 79.87% and 69.22% is repurposed such that it binds the same TF in different cells or different TFs in the same cells. Remarkably, within the set of repurposed TFos, the corresponding genome regions in the other species are preferred locations of novel TFos. These events suggest exaptation of some functional regulatory sequences into new function. Despite TFos repurposing, we did not find substantial changes in their predicted target genes, suggesting that CRMs buffer evolutionary events allowing little or no change in the TFos - target gene associations. Thus, the small portion of TFos with strictly conserved occupancy underestimates the degree of conservation of regulatory interactions.
Conclusion: We mapped regulatory sequences from an extensive number of TFs and cell types between human and mouse using WGA. A comparative analysis of this correspondence unveiled the extent of the shared regulatory sequence across TFs and cell types under study. Importantly, a large part of the shared regulatory sequence is repurposed on the other species. This sequence, fueled by turnover events, provides a strong case for exaptation in regulatory elements.
Figures
Similar articles
-
De novo prediction of cis-regulatory elements and modules through integrative analysis of a large number of ChIP datasets.BMC Genomics. 2014 Dec 2;15:1047. doi: 10.1186/1471-2164-15-1047. BMC Genomics. 2014. PMID: 25442502 Free PMC article.
-
Principles of regulatory information conservation between mouse and human.Nature. 2014 Nov 20;515(7527):371-375. doi: 10.1038/nature13985. Nature. 2014. PMID: 25409826 Free PMC article.
-
Conservation of trans-acting circuitry during mammalian regulatory evolution.Nature. 2014 Nov 20;515(7527):365-70. doi: 10.1038/nature13972. Nature. 2014. PMID: 25409825 Free PMC article.
-
Deciphering the genome's regulatory code: the many languages of DNA.Bioessays. 2010 May;32(5):381-4. doi: 10.1002/bies.200900197. Bioessays. 2010. PMID: 20394065 Free PMC article. Review.
-
Lineages, cell types and functional states: a genomic view.Curr Opin Cell Biol. 2013 Dec;25(6):759-64. doi: 10.1016/j.ceb.2013.07.006. Epub 2013 Jul 29. Curr Opin Cell Biol. 2013. PMID: 23906851 Review.
Cited by
-
Modelling the genetic aetiology of complex disease: human-mouse conservation of noncoding features and disease-associated loci.Biol Lett. 2022 Mar;18(3):20210630. doi: 10.1098/rsbl.2021.0630. Epub 2022 Mar 23. Biol Lett. 2022. PMID: 35317627 Free PMC article.
-
Evolutionary re-wiring of p63 and the epigenomic regulatory landscape in keratinocytes and its potential implications on species-specific gene expression and phenotypes.Nucleic Acids Res. 2017 Aug 21;45(14):8208-8224. doi: 10.1093/nar/gkx416. Nucleic Acids Res. 2017. PMID: 28505376 Free PMC article.
-
Discordant gene responses to radiation in humans and mice and the role of hematopoietically humanized mice in the search for radiation biomarkers.Sci Rep. 2019 Dec 19;9(1):19434. doi: 10.1038/s41598-019-55982-2. Sci Rep. 2019. PMID: 31857640 Free PMC article.
-
Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species.Proc Natl Acad Sci U S A. 2017 Dec 19;114(51):E11037-E11046. doi: 10.1073/pnas.1618075114. Epub 2017 Dec 4. Proc Natl Acad Sci U S A. 2017. PMID: 29203652 Free PMC article.
-
Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression.Nat Commun. 2015 Jan 13;6:5903. doi: 10.1038/ncomms6903. Nat Commun. 2015. PMID: 25582907 Free PMC article.
References
-
- Davidson EH. The regulatory genome: gene regulatory networks in development and evolution. Burlington (MA): Academic Press; 2006.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

