T396I mutation of mouse Sufu reduces the stability and activity of Gli3 repressor
- PMID: 25760946
- PMCID: PMC4356511
- DOI: 10.1371/journal.pone.0119455
T396I mutation of mouse Sufu reduces the stability and activity of Gli3 repressor
Abstract
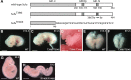
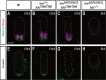
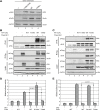
Hedgehog signaling is primarily transduced by two transcription factors: Gli2, which mainly acts as a full-length activator, and Gli3, which tends to be proteolytically processed from a full-length form (Gli3FL) to an N-terminal repressor (Gli3REP). Recent studies using a Sufu knockout mouse have indicated that Sufu is involved in regulating Gli2 and Gli3 activator and repressor activity at multiple steps of the signaling cascade; however, the mechanism of specific Gli2 and Gli3 regulation remains to be elucidated. In this study, we established an allelic series of ENU-induced mouse strains. Analysis of one of the missense alleles, SufuT396I, showed that Thr396 residue of Sufu played a key role in regulation of Gli3 activity. SufuT396I/T396I embryos exhibited severe polydactyly, which is indicative of compromised Gli3 activity. Concomitantly, significant quantitative reductions of unprocessed Gli3 (Gli3FL) and processed Gli3 (Gli3REP) were observed in vivo as well as in vitro. Genetic experiments showed that patterning defects in the limb buds of SufuT396I/T396I were rescued by a constitutive Gli3REP allele (Gli3∆699), strongly suggesting that SufuT396I reduced the truncated Gli3 repressor. In contrast, SufuT396I qualitatively exhibited no mutational effects on Gli2 regulation. Taken together, the results of this study show that the Thr396 residue of Sufu is specifically required for regulation of Gli3 but not Gli2. This implies a novel Sufu-mediated mechanism in which Gli2 activator and Gli3 repressor are differentially regulated.
Conflict of interest statement
Figures


Similar articles
-
Suppressor of Fused Is Required for Determining Digit Number and Identity via Gli3/Fgfs/Gremlin.PLoS One. 2015 May 22;10(5):e0128006. doi: 10.1371/journal.pone.0128006. eCollection 2015. PLoS One. 2015. PMID: 26001200 Free PMC article.
-
Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors.Development. 2010 Jun;137(12):2001-9. doi: 10.1242/dev.052126. Epub 2010 May 12. Development. 2010. PMID: 20463034 Free PMC article.
-
The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning.Dev Biol. 2007 May 15;305(2):460-9. doi: 10.1016/j.ydbio.2007.02.029. Epub 2007 Feb 28. Dev Biol. 2007. PMID: 17400206 Free PMC article.
-
Limb anterior-posterior polarity integrates activator and repressor functions of GLI2 as well as GLI3.Dev Biol. 2012 Oct 1;370(1):110-24. doi: 10.1016/j.ydbio.2012.07.017. Epub 2012 Jul 25. Dev Biol. 2012. PMID: 22841643 Free PMC article.
-
Suppressor of fused controls mid-hindbrain patterning and cerebellar morphogenesis via GLI3 repressor.J Neurosci. 2011 Feb 2;31(5):1825-36. doi: 10.1523/JNEUROSCI.2166-10.2011. J Neurosci. 2011. PMID: 21289193 Free PMC article.
Cited by
-
Disrupting Hedgehog signaling in melanocytes by SUFU knockout leads to ocular melanocytosis and anterior segment malformation.Dis Model Mech. 2023 Aug 1;16(8):dmm050210. doi: 10.1242/dmm.050210. Epub 2023 Aug 29. Dis Model Mech. 2023. PMID: 37577930 Free PMC article.
-
Disruption of Fuz in mouse embryos generates hypoplastic hindbrain development and reduced cranial nerve ganglia.Dev Dyn. 2024 Sep;253(9):846-858. doi: 10.1002/dvdy.702. Epub 2024 Mar 19. Dev Dyn. 2024. PMID: 38501709
-
Hypomorphic Recessive Variants in SUFU Impair the Sonic Hedgehog Pathway and Cause Joubert Syndrome with Cranio-facial and Skeletal Defects.Am J Hum Genet. 2017 Oct 5;101(4):552-563. doi: 10.1016/j.ajhg.2017.08.017. Epub 2017 Sep 28. Am J Hum Genet. 2017. PMID: 28965847 Free PMC article.
-
Congenital medulloblastoma in two brothers with SUFU-mutated Gorlin-Goltz syndrome: Case reports and literature review.Front Oncol. 2022 Oct 12;12:988798. doi: 10.3389/fonc.2022.988798. eCollection 2022. Front Oncol. 2022. PMID: 36313636 Free PMC article.
-
Suppressor of fused controls perinatal expansion and quiescence of future dentate adult neural stem cells.Elife. 2019 Apr 11;8:e42918. doi: 10.7554/eLife.42918. Elife. 2019. PMID: 30973324 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

