Improved crystallization and diffraction of caffeine-induced death suppressor protein 1 (Cid1)
- PMID: 25760713
- PMCID: PMC4356314
- DOI: 10.1107/S2053230X15001351
Improved crystallization and diffraction of caffeine-induced death suppressor protein 1 (Cid1)
Abstract
The post-transcriptional addition of uridines to the 3'-end of RNAs is an important regulatory process that is critical for coding and noncoding RNA stability. In fission yeast and metazoans this untemplated 3'-uridylylation is catalysed by a single family of terminal uridylyltransferases (TUTs) whose members are adapted to specific RNA targets. In Schizosaccharomyces pombe the TUT Cid1 is responsible for the uridylylation of polyadenylated mRNAs, targeting them for destruction. In metazoans, the Cid1 orthologues ZCCHC6 and ZCCHC11 uridylate histone mRNAs, targeting them for degradation, but also uridylate microRNAs, altering their maturation. Cid1 has been studied as a model TUT that has provided insights into the larger and more complex metazoan enzyme system. In this paper, two strategies are described that led to improvements both in the crystallogenesis of Cid1 and in the resolution of diffraction by ∼1.5 Å. These advances have allowed high-resolution crystallographic studies of this TUT system to be initiated.
Keywords: Cid1; terminal uridylyltransferases.
Figures
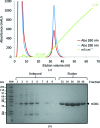
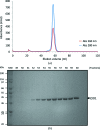


Similar articles
-
Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.Nucleic Acids Res. 2015 Mar 11;43(5):2968-79. doi: 10.1093/nar/gkv122. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712096 Free PMC article.
-
Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.Nat Struct Mol Biol. 2012 Aug;19(8):782-787. doi: 10.1038/nsmb.2329. Epub 2012 Jul 1. Nat Struct Mol Biol. 2012. PMID: 22751018 Free PMC article.
-
A critical switch in the enzymatic properties of the Cid1 protein deciphered from its product-bound crystal structure.Nucleic Acids Res. 2014 Mar;42(5):3372-80. doi: 10.1093/nar/gkt1278. Epub 2013 Dec 9. Nucleic Acids Res. 2014. PMID: 24322298 Free PMC article.
-
The Cid1 poly(U) polymerase.Biochim Biophys Acta. 2008 Apr;1779(4):286-94. doi: 10.1016/j.bbagrm.2008.03.003. Epub 2008 Mar 19. Biochim Biophys Acta. 2008. PMID: 18371314 Review.
-
The Cid1 family of non-canonical poly(A) polymerases.Yeast. 2006 Oct 15;23(13):991-1000. doi: 10.1002/yea.1408. Yeast. 2006. PMID: 17072891 Review.
Cited by
-
Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.Nucleic Acids Res. 2015 Mar 11;43(5):2968-79. doi: 10.1093/nar/gkv122. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712096 Free PMC article.
-
RNA surveillance by uridylation-dependent RNA decay in Schizosaccharomyces pombe.Nucleic Acids Res. 2019 Apr 8;47(6):3045-3057. doi: 10.1093/nar/gkz043. Nucleic Acids Res. 2019. PMID: 30715470 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

