Rapid proteasomal elimination of 3-hydroxy-3-methylglutaryl-CoA reductase by interferon-γ in primary macrophages requires endogenous 25-hydroxycholesterol synthesis
- PMID: 25759117
- PMCID: PMC4503878
- DOI: 10.1016/j.steroids.2015.02.022
Rapid proteasomal elimination of 3-hydroxy-3-methylglutaryl-CoA reductase by interferon-γ in primary macrophages requires endogenous 25-hydroxycholesterol synthesis
Abstract
Interferons (IFNs) play a central role in immunity and emerging evidence suggests that IFN-signalling coordinately regulates sterol biosynthesis in macrophages, via Sterol Regulatory Element-Binding Protein (SREBP) dependent and independent pathways. However, the precise mechanisms and kinetic steps by which IFN controls sterol biosynthesis are as yet not fully understood. Here, we elucidate the molecular circuitry governing how IFN controls the first regulated step in the mevalonate-sterol pathway, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), through the synthesis of 25-Hydroxycholesterol (25-HC) from cholesterol by the IFN-inducible Cholesterol-25-Hydroxylase (CH25H). We show for the first 30-min of IFN stimulation of macrophages the rate of de novo synthesis of the Ch25h transcript is markedly increased but by 120-min becomes transcriptionally curtailed, coincident with induction of the Activating Transcription Factor 3 (ATF3) repressor. We demonstrate ATF3 induction by Toll-like receptors is strictly dependent on IFN-signalling. While the SREBP-pathway dependent rates of de novo transcription of Hmgcr are relatively unchanged in the first 90-min of IFN treatment, we find HMGCR enzyme levels undergo a rapid proteasomal-mediated degradation, defining a previously unappreciated SREBP-independent mechanism for IFN-action. These events precede a sustained marked reduction in Hmgcr RNA levels involving SREBP-dependent mechanisms. We demonstrate that HMGCR proteasomal-degradation by IFN strictly requires the synthesis of endogenous 25-HC and functionally couples HMGCR to CH25H to coordinately suppress sterol biosynthesis. In conclusion, we quantitatively delineate proteomic and transcriptional levels of IFN-mediated control of HMGCR, the primary enzymatic step of the mevalonate-sterol biosynthesis pathway, providing a foundational framework for mathematically modelling the therapeutic outcome of immune-metabolic pathways.
Keywords: 25-Hydroxycholesterol; CH25H; Cholesterol biosynthesis; Immunity; Infection; Macrophages.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
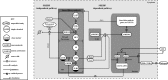
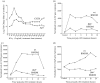

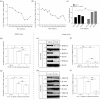

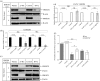
Similar articles
-
Endogenous sterol intermediates of the mevalonate pathway regulate HMGCR degradation and SREBP-2 processing.J Lipid Res. 2019 Oct;60(10):1765-1775. doi: 10.1194/jlr.RA119000201. Epub 2019 Aug 27. J Lipid Res. 2019. PMID: 31455613 Free PMC article.
-
Post-transcriptional regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase by mevalonate.Arch Biochem Biophys. 1995 Feb 20;317(1):235-43. doi: 10.1006/abbi.1995.1158. Arch Biochem Biophys. 1995. PMID: 7872789
-
Haploid Mammalian Genetic Screen Identifies UBXD8 as a Key Determinant of HMGCR Degradation and Cholesterol Biosynthesis.Arterioscler Thromb Vasc Biol. 2017 Nov;37(11):2064-2074. doi: 10.1161/ATVBAHA.117.310002. Epub 2017 Sep 7. Arterioscler Thromb Vasc Biol. 2017. PMID: 28882874 Free PMC article.
-
Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR).J Biol Chem. 2013 Jun 28;288(26):18707-15. doi: 10.1074/jbc.R113.479808. Epub 2013 May 21. J Biol Chem. 2013. PMID: 23696639 Free PMC article. Review.
-
The Potential of Isoprenoids in Adjuvant Cancer Therapy to Reduce Adverse Effects of Statins.Front Pharmacol. 2019 Jan 4;9:1515. doi: 10.3389/fphar.2018.01515. eCollection 2018. Front Pharmacol. 2019. PMID: 30662405 Free PMC article. Review.
Cited by
-
Metabolic Regulators Nampt and Sirt6 Serially Participate in the Macrophage Interferon Antiviral Cascade.Front Microbiol. 2019 Mar 4;10:355. doi: 10.3389/fmicb.2019.00355. eCollection 2019. Front Microbiol. 2019. PMID: 30886604 Free PMC article.
-
25-Hydroxycholesterol Inhibits Kaposi's Sarcoma Herpesvirus and Epstein-Barr Virus Infections and Activates Inflammatory Cytokine Responses.mBio. 2021 Dec 21;12(6):e0290721. doi: 10.1128/mBio.02907-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781692 Free PMC article.
-
The citrus flavonoid naringenin confers protection in a murine endotoxaemia model through AMPK-ATF3-dependent negative regulation of the TLR4 signalling pathway.Sci Rep. 2016 Dec 22;6:39735. doi: 10.1038/srep39735. Sci Rep. 2016. PMID: 28004841 Free PMC article.
-
Antiviral Actions of 25-Hydroxycholesterol in Fish Vary With the Virus-Host Combination.Front Immunol. 2021 Feb 24;12:581786. doi: 10.3389/fimmu.2021.581786. eCollection 2021. Front Immunol. 2021. PMID: 33717065 Free PMC article.
-
Coronavirus Infection and Cholesterol Metabolism.Front Immunol. 2022 Apr 21;13:791267. doi: 10.3389/fimmu.2022.791267. eCollection 2022. Front Immunol. 2022. PMID: 35529872 Free PMC article. Review.
References
-
- Brown M.S., Goldstein J.L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21(5):505–517. - PubMed
-
- Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425–430. - PubMed
-
- Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–340. - PubMed
-
- Goldstein J.L., DeBose-Boyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

