Regulation of CYBB Gene Expression in Human Phagocytes by a Distant Upstream NF-κB Binding Site
- PMID: 25752509
- PMCID: PMC5551681
- DOI: 10.1002/jcb.25155
Regulation of CYBB Gene Expression in Human Phagocytes by a Distant Upstream NF-κB Binding Site
Abstract
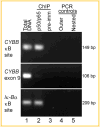
The human CYBB gene encodes the gp91-phox component of the phagocyte oxidase enzyme complex, which is responsible for generating superoxide and other downstream reactive oxygen species essential to microbial killing. In the present study, we have identified by sequence analysis a putative NF-κB binding site in a DNase I hypersensitive site, termed HS-II, located in the distant 5' flanking region of the CYBB gene. Electrophoretic mobility assays showed binding of the sequence element by recombinant NF-κB protein p50 and by proteins in nuclear extract from the HL-60 myeloid leukemia cell line corresponding to p50 and to p50/p65 heterodimers. Chromatin immunoprecipitation demonstrated NF-κB binding to the site in intact HL-60 cells. Chromosome conformation capture (3C) assays demonstrated physical interaction between the NF-κB binding site and the CYBB promoter region. Inhibition of NF-κB activity by salicylate reduced CYBB expression in peripheral blood neutrophils and differentiated U937 monocytic leukemia cells. U937 cells transfected with a mutant inhibitor of κB "super-repressor" showed markedly diminished CYBB expression. Luciferase reporter analysis of the NF-κB site linked to the CYBB 5' flanking promoter region revealed enhanced expression, augmented by treatment with interferon-γ. These studies indicate a role for this distant, 15 kb upstream, binding site in NF-κB regulation of the CYBB gene, an essential component of phagocyte-mediated host defense.
Keywords: CYBB; ENHANCER; GENE EXPRESSION; NF-κB; PHAGOCYTE OXIDASE.
© 2015 Wiley Periodicals, Inc.
Figures




Similar articles
-
Role of NF-kappaB in transcriptional regulation of the phagocyte NADPH oxidase by tumor necrosis factor-alpha.J Leukoc Biol. 2007 Sep;82(3):729-41. doi: 10.1189/jlb.1206735. Epub 2007 May 31. J Leukoc Biol. 2007. PMID: 17537988
-
JAK2 is necessary and sufficient for interferon-gamma-induced transcription of the gene encoding gp91PHOX.J Leukoc Biol. 2005 Jan;77(1):120-7. doi: 10.1189/jlb.0704429. Epub 2004 Oct 20. J Leukoc Biol. 2005. PMID: 15496449
-
Nuclear factor kappa B activation by NADPH oxidases.Mech Ageing Dev. 2004 Oct-Nov;125(10-11):799-810. doi: 10.1016/j.mad.2004.08.009. Mech Ageing Dev. 2004. PMID: 15541774
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
The identification of hematopoietic-specific regulatory elements for WASp gene expression.Mol Ther Methods Clin Dev. 2016 Dec 14;3:16077. doi: 10.1038/mtm.2016.77. eCollection 2016. Mol Ther Methods Clin Dev. 2016. PMID: 28035317 Free PMC article. Review.
Cited by
-
NOX2ko Mice Show Largely Increased Expression of a Mutated NOX2 mRNA Encoding an Inactive NOX2 Protein.Antioxidants (Basel). 2020 Oct 26;9(11):1043. doi: 10.3390/antiox9111043. Antioxidants (Basel). 2020. PMID: 33114493 Free PMC article.
-
Targeted Repair of CYBB in X-CGD iPSCs Requires Retention of Intronic Sequences for Expression and Functional Correction.Mol Ther. 2017 Feb 1;25(2):321-330. doi: 10.1016/j.ymthe.2016.11.012. Mol Ther. 2017. PMID: 28153086 Free PMC article.
-
Frontotemporal dementia: insights into the biological underpinnings of disease through gene co-expression network analysis.Mol Neurodegener. 2016 Feb 24;11:21. doi: 10.1186/s13024-016-0085-4. Mol Neurodegener. 2016. PMID: 26912063 Free PMC article.
-
A THP-1 Cell Line-Based Exploration of Immune Responses Toward Heat-Treated BLG.Front Nutr. 2021 Jan 13;7:612397. doi: 10.3389/fnut.2020.612397. eCollection 2020. Front Nutr. 2021. PMID: 33521038 Free PMC article.
-
Correction of X-CGD patient HSPCs by targeted CYBB cDNA insertion using CRISPR/Cas9 with 53BP1 inhibition for enhanced homology-directed repair.Gene Ther. 2021 Jun;28(6):373-390. doi: 10.1038/s41434-021-00251-z. Epub 2021 Mar 12. Gene Ther. 2021. PMID: 33712802 Free PMC article.
References
-
- Anrather J, Racchumi G, Iadecola C. NF-kappaB regulates phagocytic NADPH oxidase by inducing the expression of gp91phox. J Biol Chem. 2006;281:5657–5667. - PubMed
-
- Bei L, Lu Y, Bellis SL, Zhou W, Horvath E, Eklund EA. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding beta3 integrin during myeloid differentiation. J Biol Chem. 2007;282:16846–16859. page-First>. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

