Spatial perturbation with synthetic protein scaffold reveals robustness of asymmetric cell division
- PMID: 25750689
- PMCID: PMC4350780
- DOI: 10.4236/jbise.2013.62017
Spatial perturbation with synthetic protein scaffold reveals robustness of asymmetric cell division
Abstract
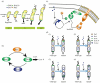
Asymmetric cell division is an important mechanism for creating diversity in a cellular population. Stem cells commonly perform asymmetric division to generate both a daughter stem cell for self-renewal and a more differentiated daughter cell to populate the tissue. During asymmetric cell division, protein cell fate determinants asymmetrically localize to the opposite poles of a dividing cell to cause distinct cell fate. However, it remains unclear whether cell fate determination is robust to fluctuations and noise during this spatial allocation process. To answer this question, we engineered Caulobacter, a bacterial model for asymmetric division, to express synthetic scaffolds with modular protein interaction domains. These scaffolds perturbed the spatial distribution of the PleC-DivJ-DivK phospho-signaling network without changing their endogenous expression levels. Surprisingly, enforcing symmetrical distribution of these cell fate determinants did not result in symmetric daughter fate or any morphological defects. Further computational analysis suggested that PleC and DivJ form a robust phospho-switch that can tolerate high amount of spatial variation. This insight may shed light on the presence of similar phospho-switches in stem cell asymmetric division regulation. Overall, our study demonstrates that synthetic protein scaffolds can provide a useful tool to probe biological systems for better understanding of their operating principles.
Keywords: Asymmetric Cell Division; Caulobacter; Protein Scaffold; Synthetic Biology.
Figures



Similar articles
-
The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus.Genes Dev. 2008 Jan 15;22(2):212-25. doi: 10.1101/gad.1601808. Genes Dev. 2008. PMID: 18198338 Free PMC article.
-
Interplay between the localization and kinetics of phosphorylation in flagellar pole development of the bacterium Caulobacter crescentus.PLoS Comput Biol. 2012;8(8):e1002602. doi: 10.1371/journal.pcbi.1002602. Epub 2012 Aug 2. PLoS Comput Biol. 2012. PMID: 22876167 Free PMC article.
-
Dynamical Localization of DivL and PleC in the Asymmetric Division Cycle of Caulobacter crescentus: A Theoretical Investigation of Alternative Models.PLoS Comput Biol. 2015 Jul 17;11(7):e1004348. doi: 10.1371/journal.pcbi.1004348. eCollection 2015 Jul. PLoS Comput Biol. 2015. PMID: 26186202 Free PMC article.
-
The asymmetric division and tumorigenesis of stem cells.Chin J Cancer. 2010 Mar;29(3):248-53. doi: 10.5732/cjc.009.10668. Chin J Cancer. 2010. PMID: 20193105 Review.
-
Asymmetric cell division of mammary stem cells.Cell Div. 2021 Sep 29;16(1):5. doi: 10.1186/s13008-021-00073-w. Cell Div. 2021. PMID: 34587981 Free PMC article. Review.
References
-
- Tajbakhsh S, Rocheteau P, Le Roux I. Asymmetric cell divisions and asymmetric cell fates. Annual Review of Cell and Developmental Biology. 2009;25:671–699. doi:10.1146/annurev.cellbio.24.110707.175415. - PubMed
-
- Knoblich JA. Asymmetric cell division during animal development. Nature Reviews. Molecular Cell Biology. 2001;2:11–20. doi:10.1038/35048085. - PubMed
-
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi:10.1038/nature04956. - PubMed
-
- Caussinus E, Hirth F. Asymmetric stem cell division in development and cancer. Progress in Molecular and Subcellular Biology. 2007;45:205–225. doi:10.1007/978-3-540-69161-7_9. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
