Toward optimization of AgoshRNA molecules that use a non-canonical RNAi pathway: variations in the top and bottom base pairs
- PMID: 25747107
- PMCID: PMC4615845
- DOI: 10.1080/15476286.2015.1022024
Toward optimization of AgoshRNA molecules that use a non-canonical RNAi pathway: variations in the top and bottom base pairs
Abstract
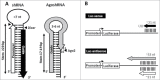
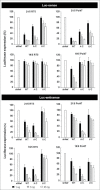
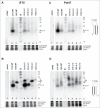
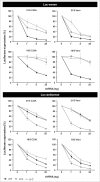
Short hairpin RNAs (shRNAs) are widely used for gene knockdown by inducing the RNA interference (RNAi) mechanism. The shRNA precursor is processed by Dicer into small interfering RNAs (siRNAs) and subsequently programs the RNAi-induced silencing complex (RISC) to find a complementary target mRNA (mRNA) for post-transcriptional gene silencing. Recent evidence indicates that shRNAs with a relatively short basepaired stem bypass Dicer to be processed directly by the Ago2 nuclease of the RISC complex. We named this design AgoshRNA as these molecules depend on Ago2 both for processing and subsequent silencing activity. This alternative AgoshRNA processing route yields only a single active RNA strand, an important feature to restrict off-target effects induced by the passenger strand of regular shRNAs. It is therefore important to understand this novel AgoshRNA processing route in mechanistic detail such that one can design the most effective and selective RNA reagents. We performed a systematic analysis of the optimal base pair (bp) composition at the top and bottom of AgoshRNA molecules. In this study, we document the importance of the 5' end nucleotide (nt) and a bottom mismatch. The optimized AgoshRNA design exhibits improved RNAi activity across cell types. These results have important implications for the future design of more specific RNAi therapeutics.
Keywords: RNA processing; agoshRNA; argonaute2; dicer; shRNA.
Figures

Similar articles
-
Influence of the loop size and nucleotide composition on AgoshRNA biogenesis and activity.RNA Biol. 2017 Nov 2;14(11):1559-1569. doi: 10.1080/15476286.2017.1328349. Epub 2017 Nov 3. RNA Biol. 2017. PMID: 28569591 Free PMC article.
-
Mechanistic insights on the Dicer-independent AGO2-mediated processing of AgoshRNAs.RNA Biol. 2015;12(1):92-100. doi: 10.1080/15476286.2015.1017204. RNA Biol. 2015. PMID: 25826416 Free PMC article.
-
The influence of the 5΄-terminal nucleotide on AgoshRNA activity and biogenesis: importance of the polymerase III transcription initiation site.Nucleic Acids Res. 2017 Apr 20;45(7):4036-4050. doi: 10.1093/nar/gkw1203. Nucleic Acids Res. 2017. PMID: 27928054 Free PMC article.
-
Approaches for chemically synthesized siRNA and vector-mediated RNAi.FEBS Lett. 2005 Oct 31;579(26):5974-81. doi: 10.1016/j.febslet.2005.08.070. Epub 2005 Sep 20. FEBS Lett. 2005. PMID: 16199038 Review.
-
Technology of RNA Interference in Advanced Medicine.Microrna. 2018;7(2):74-84. doi: 10.2174/2211536607666180129153307. Microrna. 2018. PMID: 29380708 Review.
Cited by
-
Mutation of nucleotides around the +1 position of type 3 polymerase III promoters: The effect on transcriptional activity and start site usage.Transcription. 2017;8(5):275-287. doi: 10.1080/21541264.2017.1322170. Epub 2017 Jun 9. Transcription. 2017. PMID: 28598252 Free PMC article.
-
Novel AgoshRNA molecules for silencing of the CCR5 co-receptor for HIV-1 infection.PLoS One. 2017 May 24;12(5):e0177935. doi: 10.1371/journal.pone.0177935. eCollection 2017. PLoS One. 2017. PMID: 28542329 Free PMC article.
-
Bone Marrow Gene Therapy for HIV/AIDS.Viruses. 2015 Jul 17;7(7):3910-36. doi: 10.3390/v7072804. Viruses. 2015. PMID: 26193303 Free PMC article. Review.
-
Influence of a 3' Terminal Ribozyme on AgoshRNA Biogenesis and Activity.Mol Ther Nucleic Acids. 2019 Jun 7;16:452-462. doi: 10.1016/j.omtn.2019.04.001. Epub 2019 Apr 8. Mol Ther Nucleic Acids. 2019. PMID: 31048184 Free PMC article.
-
Design of antiviral AGO2-dependent short hairpin RNAs.Virol Sin. 2024 Aug;39(4):645-654. doi: 10.1016/j.virs.2024.05.001. Epub 2024 May 9. Virol Sin. 2024. PMID: 38734183 Free PMC article.
References
-
- Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 2012; 40:4626-40; PMID:22270084; http://dx.doi.org/10.1093/nar/gks026 - DOI - PMC - PubMed
-
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43:892-903; PMID:21925378; http://dx.doi.org/10.1016/j.molcel.2011.07.024 - DOI - PMC - PubMed
-
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature 2010; 465:584-9; PMID:20424607; http://dx.doi.org/10.1038/nature09092 - DOI - PMC - PubMed
-
- Berkhout B, Liu YP. Towards improved shRNA and miRNA reagents as inhibitors of HIV-1 replication. Future Microbiol 2014; 9:561-71; PMID:24810353; http://dx.doi.org/10.2217/fmb.14.5 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
