Multispecies Analysis of Expression Pattern Diversification in the Recently Expanded Insect Ly6 Gene Family
- PMID: 25743545
- PMCID: PMC4476152
- DOI: 10.1093/molbev/msv052
Multispecies Analysis of Expression Pattern Diversification in the Recently Expanded Insect Ly6 Gene Family
Abstract
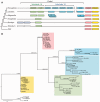
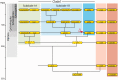
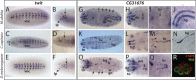
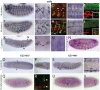
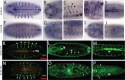
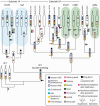
Gene families often consist of members with diverse expression domains reflecting their functions in a wide variety of tissues. However, how the expression of individual members, and thus their tissue-specific functions, diversified during the course of gene family expansion is not well understood. In this study, we approached this question through the analysis of the duplication history and transcriptional evolution of a rapidly expanding subfamily of insect Ly6 genes. We analyzed different insect genomes and identified seven Ly6 genes that have originated from a single ancestor through sequential duplication within the higher Diptera. We then determined how the original embryonic expression pattern of the founding gene diversified by characterizing its tissue-specific expression in the beetle Tribolium castaneum, the butterfly Bicyclus anynana, and the mosquito Anopheles stephensi and those of its duplicates in three higher dipteran species, representing various stages of the duplication history (Megaselia abdita, Ceratitis capitata, and Drosophila melanogaster). Our results revealed that frequent neofunctionalization episodes contributed to the increased expression breadth of this subfamily and that these events occurred after duplication and speciation events at comparable frequencies. In addition, at each duplication node, we consistently found asymmetric expression divergence. One paralog inherited most of the tissue-specificities of the founder gene, whereas the other paralog evolved drastically reduced expression domains. Our approach attests to the power of combining a well-established duplication history with a comprehensive coverage of representative species in acquiring unequivocal information about the dynamics of gene expression evolution in gene families.
Keywords: Drosophila; Ly6 protein; gene duplication; gene family evolution; regulatory evolution.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Comparative genomic analysis and evolution of family-B G protein-coupled receptors from six model insect species.Gene. 2013 Apr 25;519(1):1-12. doi: 10.1016/j.gene.2013.01.061. Epub 2013 Feb 18. Gene. 2013. PMID: 23428791
-
Diverse Cis-Regulatory Mechanisms Contribute to Expression Evolution of Tandem Gene Duplicates.Mol Biol Evol. 2017 Dec 1;34(12):3132-3147. doi: 10.1093/molbev/msx237. Mol Biol Evol. 2017. PMID: 28961967 Free PMC article.
-
Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation.Insect Biochem Mol Biol. 2007 Aug;37(8):741-53. doi: 10.1016/j.ibmb.2007.02.012. Epub 2007 Mar 5. Insect Biochem Mol Biol. 2007. PMID: 17628274
-
A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum.Front Neuroendocrinol. 2008 Jan;29(1):142-65. doi: 10.1016/j.yfrne.2007.10.003. Epub 2007 Oct 24. Front Neuroendocrinol. 2008. PMID: 18054377 Review.
-
The evolution of developmental gene networks: lessons from comparative studies on holometabolous insects.Philos Trans R Soc Lond B Biol Sci. 2008 Apr 27;363(1496):1539-47. doi: 10.1098/rstb.2007.2244. Philos Trans R Soc Lond B Biol Sci. 2008. PMID: 18192180 Free PMC article. Review.
Cited by
-
Diversification of spatiotemporal expression and copy number variation of the echinoid hbox12/pmar1/micro1 multigene family.PLoS One. 2017 Mar 28;12(3):e0174404. doi: 10.1371/journal.pone.0174404. eCollection 2017. PLoS One. 2017. PMID: 28350855 Free PMC article.
-
Comparative genomic analysis of SET domain family reveals the origin, expansion, and putative function of the arthropod-specific SmydA genes as histone modifiers in insects.Gigascience. 2017 Jun 1;6(6):1-16. doi: 10.1093/gigascience/gix031. Gigascience. 2017. PMID: 28444351 Free PMC article.
-
A distinct Acyl-CoA binding protein (ACBP6) shapes tissue plasticity during nutrient adaptation in Drosophila.Nat Commun. 2023 Nov 21;14(1):7599. doi: 10.1038/s41467-023-43362-4. Nat Commun. 2023. PMID: 37989752 Free PMC article.
-
The number of neurons in Drosophila and mosquito brains.PLoS One. 2021 May 14;16(5):e0250381. doi: 10.1371/journal.pone.0250381. eCollection 2021. PLoS One. 2021. PMID: 33989293 Free PMC article.
-
Parasitoid wasp venom vesicles (venosomes) enter Drosophila melanogaster lamellocytes through a flotillin/lipid raft-dependent endocytic pathway.Virulence. 2020 Dec;11(1):1512-1521. doi: 10.1080/21505594.2020.1838116. Virulence. 2020. PMID: 33135553 Free PMC article.
References
-
- Alonso CR, Wilkins AS. The molecular elements that underlie developmental evolution. Nat Rev Genet. 2005;6:709–715. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

