Transmitted/Founder Viruses Rapidly Escape from CD8+ T Cell Responses in Acute Hepatitis C Virus Infection
- PMID: 25740982
- PMCID: PMC4442533
- DOI: 10.1128/JVI.03717-14
Transmitted/Founder Viruses Rapidly Escape from CD8+ T Cell Responses in Acute Hepatitis C Virus Infection
Abstract
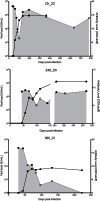
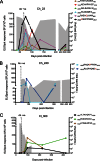
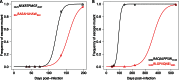
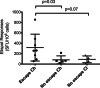
The interaction between hepatitis C virus (HCV) and cellular immune responses during very early infection is critical for disease outcome. To date, the impact of antigen-specific cellular immune responses on the evolution of the viral population establishing infection and on potential escape has not been studied. Understanding these early host-virus dynamics is important for the development of a preventative vaccine. Three subjects who were followed longitudinally from the detection of viremia preseroconversion until disease outcome were analyzed. The evolution of transmitted/founder (T/F) viruses was undertaken using deep sequencing. CD8(+) T cell responses were measured via enzyme-linked immunosorbent spot (ELISpot) assay using HLA class I-restricted T/F epitopes. T/F viruses were rapidly extinguished in all subjects associated with either viral clearance (n = 1) or replacement with viral variants leading to establishment of chronic infection (n = 2). CD8(+) T cell responses against 11 T/F epitopes were detectable by 33 to 44 days postinfection, and 5 of these epitopes had not previously been reported. These responses declined rapidly in those who became chronically infected and were maintained in the subject who cleared infection. Higher-magnitude CD8(+) T cell responses were associated with rapid development of immune escape variants at a rate of up to 0.1 per day. Rapid escape from CD8(+) T cell responses has been quantified for the first time in the early phase of primary HCV infection. These rapid escape dynamics were associated with higher-magnitude CD8(+) T cell responses. These findings raise questions regarding optimal selection of immunogens for HCV vaccine development and suggest that detailed analysis of individual epitopes may be required.
Importance: A major limitation in our detailed understanding of the role of immune response in HCV clearance has been the lack of data on very early primary infection when the transmitted viral variants successfully establish the acute infection. This study was made possible through the availability of specimens from a unique cohort of asymptomatic primary infection cases in whom the first available viremic samples were collected approximately 3 weeks postinfection and at regular intervals thereafter. The study included detailed examination of both the evolution of the viral population and the host cellular immune responses against the T/F viruses. The findings here provide the first evidence of host cellular responses targeting T/F variants and imposing a strong selective force toward viral escape. The results of this study provide useful insight on how virus escapes the host response and consequently on future analysis of vaccine-induced immunity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Positive selection of cytotoxic T lymphocyte escape variants during acute hepatitis C virus infection.Eur J Immunol. 2005 Sep;35(9):2627-37. doi: 10.1002/eji.200526067. Eur J Immunol. 2005. PMID: 16114108
-
Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution.Hepatology. 2006 Mar;43(3):563-72. doi: 10.1002/hep.21049. Hepatology. 2006. PMID: 16496339
-
CD8 epitope escape and reversion in acute HCV infection.J Exp Med. 2004 Dec 20;200(12):1593-604. doi: 10.1084/jem.20041006. J Exp Med. 2004. PMID: 15611288 Free PMC article.
-
Hepatitis C virus--T-cell responses and viral escape mutations.Eur J Immunol. 2012 Jan;42(1):17-26. doi: 10.1002/eji.201141593. Epub 2011 Nov 28. Eur J Immunol. 2012. PMID: 22125159 Review.
-
Success and failure of virus-specific T cell responses in hepatitis C virus infection.Dig Dis. 2011;29(4):416-22. doi: 10.1159/000329807. Epub 2011 Aug 30. Dig Dis. 2011. PMID: 21894013 Review.
Cited by
-
Genetic Diversity Underlying the Envelope Glycoproteins of Hepatitis C Virus: Structural and Functional Consequences and the Implications for Vaccine Design.Viruses. 2015 Jul 17;7(7):3995-4046. doi: 10.3390/v7072809. Viruses. 2015. PMID: 26193307 Free PMC article. Review.
-
Update on Hepatitis C Vaccine: Results and Challenges.Viruses. 2024 Aug 21;16(8):1337. doi: 10.3390/v16081337. Viruses. 2024. PMID: 39205311 Free PMC article. Review.
-
Identification of human progenitors of exhausted CD8+ T cells associated with elevated IFN-γ response in early phase of viral infection.Nat Commun. 2022 Dec 7;13(1):7543. doi: 10.1038/s41467-022-35281-7. Nat Commun. 2022. PMID: 36477661 Free PMC article.
-
Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells.Immunol Cell Biol. 2016 Jul;94(6):604-11. doi: 10.1038/icb.2016.16. Epub 2016 Feb 10. Immunol Cell Biol. 2016. PMID: 26860370
-
Cytomegalovirus-Specific T Cell Epitope Recognition in Congenital Cytomegalovirus Mother-Infant Pairs.Front Immunol. 2020 Nov 24;11:568217. doi: 10.3389/fimmu.2020.568217. eCollection 2020. Front Immunol. 2020. PMID: 33329532 Free PMC article.
References
-
- Grebely J, Page K, Sacks-Davis R, van der Loeff MS, Rice TM, Bruneau J, Morris MD, Hajarizadeh B, Amin J, Cox AL, Kim AY, McGovern BH, Schinkel J, George J, Shoukry NH, Lauer GM, Maher L, Lloyd AR, Hellard M, Dore GJ, Prins M, InC3 Study Group. 2014. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology 59:109–120. doi:10.1002/hep.26639. - DOI - PMC - PubMed
-
- Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, Learn GH, Hraber P, Goepfert PA, Saag MS, Denny TN, Haynes BF, Hahn BH, Ribeiro RM, Perelson AS, Korber BT, Bhattacharya T, Shaw GM. 2012. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog 8:e1002880. doi:10.1371/journal.ppat.1002880. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

