Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma
- PMID: 25735706
- PMCID: PMC4348413
- DOI: 10.3402/jev.v4.26659
Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma
Abstract
Background: Lung cancer is one of the leading causes of cancer-related death. At the time of diagnosis, more than half of the patients will have disseminated disease and, yet, diagnosing can be challenging. New methods are desired to improve the diagnostic work-up. Exosomes are cell-derived vesicles displaying various proteins on their membrane surfaces. In addition, they are readily available in blood samples where they constitute potential biomarkers of human diseases, such as cancer. Here, we examine the potential of distinguishing non-small cell lung carcinoma (NSCLC) patients from control subjects based on the differential display of exosomal protein markers.
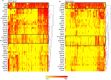
Methods: Plasma was isolated from 109 NSCLC patients with advanced stage (IIIa-IV) disease and 110 matched control subjects initially suspected of having cancer, but diagnosed to be cancer free. The Extracellular Vesicle Array (EV Array) was used to phenotype exosomes directly from the plasma samples. The array contained 37 antibodies targeting lung cancer-related proteins and was used to capture exosomes, which were visualised with a cocktail of biotin-conjugated CD9, CD63 and CD81 antibodies.
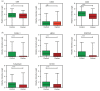
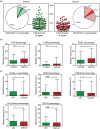
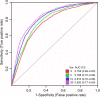
Results: The EV Array analysis was capable of detecting and phenotyping exosomes in all samples from only 10 µL of unpurified plasma. Multivariate analysis using the Random Forests method produced a combined 30-marker model separating the two patient groups with an area under the curve of 0.83, CI: 0.77-0.90. The 30-marker model has a sensitivity of 0.75 and a specificity of 0.76, and it classifies patients with 75.3% accuracy.
Conclusion: The EV Array technique is a simple, minimal-invasive tool with potential to identify lung cancer patients.
Keywords: EV Array; NSCLC; exosomes; extracellular vesicles; lung cancer; phenotyping; plasma; protein microarray.
Figures
Similar articles
-
Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping.J Extracell Vesicles. 2013 Jun 18;2. doi: 10.3402/jev.v2i0.20920. eCollection 2013. J Extracell Vesicles. 2013. PMID: 24009888 Free PMC article.
-
Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer.J Thorac Oncol. 2016 Oct;11(10):1701-10. doi: 10.1016/j.jtho.2016.05.034. Epub 2016 Jun 22. J Thorac Oncol. 2016. PMID: 27343445
-
Exosomal proteins as prognostic biomarkers in non-small cell lung cancer.Mol Oncol. 2016 Dec;10(10):1595-1602. doi: 10.1016/j.molonc.2016.10.003. Epub 2016 Oct 21. Mol Oncol. 2016. PMID: 27856179 Free PMC article.
-
Exosome beads array for multiplexed phenotyping in cancer.J Proteomics. 2019 Apr 30;198:87-97. doi: 10.1016/j.jprot.2018.12.023. Epub 2018 Dec 28. J Proteomics. 2019. PMID: 30594577 Review.
-
Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers.Asia Pac J Clin Oncol. 2018 Dec;14(6):383-391. doi: 10.1111/ajco.12869. Epub 2018 Mar 25. Asia Pac J Clin Oncol. 2018. PMID: 29575602 Review.
Cited by
-
Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples.J Extracell Vesicles. 2016 Apr 15;5:25355. doi: 10.3402/jev.v5.25355. eCollection 2016. J Extracell Vesicles. 2016. PMID: 27086589 Free PMC article.
-
Comparison and optimization of nanoscale extracellular vesicle imaging by scanning electron microscopy for accurate size-based profiling and morphological analysis.Nanoscale Adv. 2021 Mar 22;3(11):3053-3063. doi: 10.1039/d0na00948b. eCollection 2021 Jun 1. Nanoscale Adv. 2021. PMID: 36133670 Free PMC article.
-
Personalized medicine: From diagnostic to adaptive.Biomed J. 2022 Feb;45(1):132-142. doi: 10.1016/j.bj.2019.05.004. Epub 2019 Jul 9. Biomed J. 2022. PMID: 35590431 Free PMC article. Review.
-
Comparative proteomic profiling of plasma exosomes in lung cancer cases of liver and brain metastasis.Cell Biosci. 2023 Sep 28;13(1):180. doi: 10.1186/s13578-023-01112-5. Cell Biosci. 2023. PMID: 37770976 Free PMC article.
-
Multiplexed immunophenotyping of circulating exosomes on nano-engineered ExoProfile chip towards early diagnosis of cancer.Chem Sci. 2019 Apr 22;10(21):5495-5504. doi: 10.1039/c9sc00961b. eCollection 2019 Jun 7. Chem Sci. 2019. PMID: 31293733 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. - PubMed
-
- American Cancer Society. Cancer facts and figures 2014 [Internet] 2014. [cited 2014 Sep 22]. Available from: http://www.cancer.org/acs/groups/content/@research/documents/webcontent/....
-
- Engholm G, Ferlay J, Christensen N, Kejs AMT, Johannesen TB, Khan S, et al. NORDCAN: Cancer Incidence, Mortality, Prevalence and Survival in the Nordic Countries, Version 6.0. Association of the Nordic Cancer Registries. Danish Cancer Society. 2011. [cited 2014 Sep 22]. Available from: http://www.ancr.nu.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

