Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture
- PMID: 25732821
- PMCID: PMC4542312
- DOI: 10.1016/j.celrep.2015.02.004
Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture
Abstract
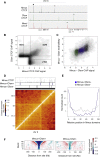
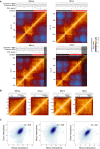
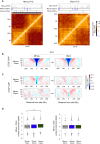
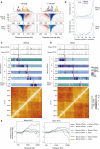
Topological domains are key architectural building blocks of chromosomes, but their functional importance and evolutionary dynamics are not well defined. We performed comparative high-throughput chromosome conformation capture (Hi-C) in four mammals and characterized the conservation and divergence of chromosomal contact insulation and the resulting domain architectures within distantly related genomes. We show that the modular organization of chromosomes is robustly conserved in syntenic regions and that this is compatible with conservation of the binding landscape of the insulator protein CTCF. Specifically, conserved CTCF sites are co-localized with cohesin, are enriched at strong topological domain borders, and bind to DNA motifs with orientations that define the directionality of CTCF's long-range interactions. Conversely, divergent CTCF binding between species is correlated with divergence of internal domain structure, likely driven by local CTCF binding sequence changes, demonstrating how genome evolution can be linked to a continuous flux of local conformation changes. We also show that large-scale domains are reorganized during genome evolution as intact modules.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Genetic Tailors: CTCF and Cohesin Shape the Genome During Evolution.Trends Genet. 2015 Nov;31(11):651-660. doi: 10.1016/j.tig.2015.09.004. Epub 2015 Oct 1. Trends Genet. 2015. PMID: 26439501 Review.
-
Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders.Genome Biol. 2016 Aug 31;17(1):182. doi: 10.1186/s13059-016-1043-8. Genome Biol. 2016. PMID: 27582050 Free PMC article.
-
Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions.Genome Res. 2013 Oct;23(10):1624-35. doi: 10.1101/gr.150136.112. Epub 2013 Jun 26. Genome Res. 2013. PMID: 23804403 Free PMC article.
-
Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells.Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):996-1001. doi: 10.1073/pnas.1317788111. Epub 2013 Dec 13. Proc Natl Acad Sci U S A. 2014. PMID: 24335803 Free PMC article.
-
Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation.J Biol Chem. 2012 Sep 7;287(37):30906-13. doi: 10.1074/jbc.R111.324962. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952237 Free PMC article. Review.
Cited by
-
CTCF As an Example of DNA-Binding Transcription Factors Containing Clusters of C2H2-Type Zinc Fingers.Acta Naturae. 2021 Jan-Mar;13(1):31-46. doi: 10.32607/actanaturae.11206. Acta Naturae. 2021. PMID: 33959385 Free PMC article.
-
Transcriptional Dynamics at Brain Enhancers: from Functional Specialization to Neurodegeneration.Curr Neurol Neurosci Rep. 2016 Oct;16(10):94. doi: 10.1007/s11910-016-0689-7. Curr Neurol Neurosci Rep. 2016. PMID: 27628759 Free PMC article. Review.
-
Characterizing Genetic Regulatory Elements in Ovine Tissues.Front Genet. 2021 May 20;12:628849. doi: 10.3389/fgene.2021.628849. eCollection 2021. Front Genet. 2021. PMID: 34093640 Free PMC article.
-
Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations.Genome Res. 2016 Jun;26(6):719-31. doi: 10.1101/gr.201517.115. Epub 2016 Apr 6. Genome Res. 2016. PMID: 27053337 Free PMC article.
-
A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires.Nat Immunol. 2015 Oct;16(10):1085-93. doi: 10.1038/ni.3232. Epub 2015 Aug 10. Nat Immunol. 2015. PMID: 26258942 Free PMC article.
References
-
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
-
- Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
-
- Borneman A.R., Gianoulis T.A., Zhang Z.D., Yu H., Rozowsky J., Seringhaus M.R., Wang L.Y., Gerstein M., Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

