Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots
- PMID: 25728785
- PMCID: PMC4692092
- DOI: 10.1002/pmic.201400530
Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots
Abstract
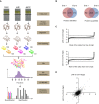
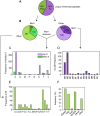
According to the Arg/N-end rule pathway, proteins with basic N-termini are targeted for degradation by the Arabidopsis thaliana E3 ligase, PROTEOLYSIS6 (PRT6). Proteins can also become PRT6 substrates following post-translational arginylation by arginyltransferases ATE1 and 2. Here, we undertook a quantitative proteomics study of Arg/N-end rule mutants, ate1/2 and prt6, to investigate the impact of this pathway on the root proteome. Tandem mass tag labelling identified a small number of proteins with increased abundance in the mutants, some of which represent downstream targets of transcription factors known to be N-end rule substrates. Isolation of N-terminal peptides using terminal amine isotope labelling of samples (TAILS) combined with triple dimethyl labelling identified 1465 unique N-termini. Stabilising residues were over-represented among the free neo-N-termini, but destabilising residues were not markedly enriched in N-end rule mutants. The majority of free neo-N-termini were revealed following cleavage of organellar targeting signals, thus compartmentation may account in part for the presence of destabilising residues in the wild-type N-terminome. Our data suggest that PRT6 does not have a marked impact on the global proteome of Arabidopsis roots and is likely involved in the controlled degradation of relatively few regulatory proteins. All MS data have been deposited in the ProteomeXchange with identifier PXD001719 (http://proteomecentral.proteomexchange.org/dataset/PXD001719).
Keywords: N-end rule; Plant proteomics; Quantitative proteomics; Root; TAILS; TMT.
© 2015 The Authors. PROTEOMICS published by Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


Similar articles
-
N-terminomics reveals control of Arabidopsis seed storage proteins and proteases by the Arg/N-end rule pathway.New Phytol. 2018 May;218(3):1106-1126. doi: 10.1111/nph.14909. Epub 2017 Nov 23. New Phytol. 2018. PMID: 29168982 Free PMC article.
-
PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus.FEBS Lett. 2007 Jul 10;581(17):3189-96. doi: 10.1016/j.febslet.2007.06.005. Epub 2007 Jun 12. FEBS Lett. 2007. PMID: 17572409
-
An improved workflow for quantitative N-terminal charge-based fractional diagonal chromatography (ChaFRADIC) to study proteolytic events in Arabidopsis thaliana.Proteomics. 2015 Jul;15(14):2458-69. doi: 10.1002/pmic.201500014. Proteomics. 2015. PMID: 26010716
-
Physiological functions and clinical implications of the N-end rule pathway.Front Med. 2016 Sep;10(3):258-70. doi: 10.1007/s11684-016-0458-7. Epub 2016 Sep 7. Front Med. 2016. PMID: 27492620 Review.
-
Post-translational N-terminal Arginylation of Protein Fragments: A Pivotal Portal to Proteolysis.Curr Protein Pept Sci. 2018;19(12):1214-1223. doi: 10.2174/1389203719666180809113122. Curr Protein Pept Sci. 2018. PMID: 30091410 Review.
Cited by
-
Protein Processing in Plant Mitochondria Compared to Yeast and Mammals.Front Plant Sci. 2022 Feb 2;13:824080. doi: 10.3389/fpls.2022.824080. eCollection 2022. Front Plant Sci. 2022. PMID: 35185991 Free PMC article. Review.
-
The Arabidopsis PeptideAtlas: Harnessing worldwide proteomics data to create a comprehensive community proteomics resource.Plant Cell. 2021 Nov 4;33(11):3421-3453. doi: 10.1093/plcell/koab211. Plant Cell. 2021. PMID: 34411258 Free PMC article.
-
Label-Free Quantitative Proteomics Reveal the Involvement of PRT6 in Arabidopsis thaliana Seed Responsiveness to Ethylene.Int J Mol Sci. 2022 Aug 19;23(16):9352. doi: 10.3390/ijms23169352. Int J Mol Sci. 2022. PMID: 36012613 Free PMC article.
-
Ethylene augments root hypoxia tolerance via growth cessation and reactive oxygen species amelioration.Plant Physiol. 2022 Sep 28;190(2):1365-1383. doi: 10.1093/plphys/kiac245. Plant Physiol. 2022. PMID: 35640551 Free PMC article.
-
Analyzing N-terminal Arginylation through the Use of Peptide Arrays and Degradation Assays.J Biol Chem. 2016 Sep 30;291(40):20976-20992. doi: 10.1074/jbc.M116.747956. Epub 2016 Aug 10. J Biol Chem. 2016. PMID: 27510035 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

