Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic
- PMID: 25724305
- PMCID: PMC4368840
- DOI: 10.1007/s10549-015-3313-7
Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic
Abstract
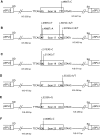
Pathogenic germline mutations in the BRCA1 gene predispose carriers to early onset breast and ovarian cancer. Clinical genetic screening of BRCA1 often reveals variants with uncertain clinical significance, complicating patient and family management. Therefore, functional examinations are urgently needed to classify whether these uncertain variants are pathogenic or benign. In this study, we investigated 14 BRCA1 variants by in silico splicing analysis and mini-gene splicing assay. All 14 alterations were missense variants located within the BRCT domain of BRCA1 and had previously been examined by functional analysis at the protein level. Results from a validated mini-gene splicing assay indicated that nine BRCA1 variants resulted in splicing aberrations leading to truncated transcripts and thus can be considered pathogenic (c.4987A>T/p.Met1663Leu, c.4988T>A/p.Met1663Lys, c.5072C>T/p.Thr1691Ile, c.5074G>C/p.Asp1692His, c.5074G>A/p.Asp1692Asn, c.5074G>T/p.Asp1692Tyr, c.5332G>A/p.Asp1778Asn, c.5332G>T/p.Asp1778Tyr, and c.5408G>C/p.Gly1803Ala), whereas five BRCA1 variants had no effect on splicing (c.4985T>C/p.Phe1662Ser, c.5072C>A/p.Thr1691Lys, c.5153G>C/p.Trp1718Ser, c.5154G>T/p.Trp1718Cys, and c.5333A>G/p.Asp1778Gly). Eight of the variants having an effect on splicing (c.4987A>T/p.Met1663Leu, c.4988T>A/p.Met1663Lys, c.5074G>C/p.Asp1692His, c.5074G>A/p.Asp1692Asn, c.5074G>T/p.Asp1692Tyr, c.5332G>A/p.Asp1778Asn, c.5332G>T/p.Asp1778Tyr, and c.5408G>C/p.Gly1803Ala) were previously determined to have no or an uncertain effect on the protein level, whereas one variant (c.5072C>T/p.Thr1691Ile) were shown to have a strong effect on the protein level as well. In conclusion, our study emphasizes that in silico splicing prediction and mini-gene splicing analysis are important for the classification of BRCA1 missense variants located close to exon/intron boundaries.
Figures

Similar articles
-
Characterization of BRCA1 and BRCA2 splicing variants: a collaborative report by ENIGMA consortium members.Breast Cancer Res Treat. 2012 Apr;132(3):1009-23. doi: 10.1007/s10549-011-1674-0. Epub 2011 Jul 19. Breast Cancer Res Treat. 2012. PMID: 21769658
-
Additional molecular and clinical evidence open the way to definitive IARC classification of the BRCA1 c.5332G > A (p.Asp1778Asn) variant.Clin Biochem. 2019 Jan;63:54-58. doi: 10.1016/j.clinbiochem.2018.10.004. Epub 2018 Oct 10. Clin Biochem. 2019. PMID: 30315757
-
Functional characterization of BRCA1 gene variants by mini-gene splicing assay.Eur J Hum Genet. 2014 Dec;22(12):1362-8. doi: 10.1038/ejhg.2014.40. Epub 2014 Mar 26. Eur J Hum Genet. 2014. PMID: 24667779 Free PMC article.
-
Understanding germ-line mutations in BRCA1.Cancer Biol Ther. 2004 Jun;3(6):515-20. doi: 10.4161/cbt.3.6.841. Epub 2004 Jun 1. Cancer Biol Ther. 2004. PMID: 15254424 Review.
-
Aberrant RNA splicing as the molecular basis of some pathogenic variants.Yi Chuan. 2017 Mar 20;39(3):200-207. doi: 10.16288/j.yczz.16-336. Yi Chuan. 2017. PMID: 28420616 Review.
Cited by
-
Low-level constitutional mosaicism of BRCA1 in two women with young onset ovarian cancer.Hered Cancer Clin Pract. 2022 Sep 6;20(1):32. doi: 10.1186/s13053-022-00237-x. Hered Cancer Clin Pract. 2022. PMID: 36068545 Free PMC article.
-
Genetic screening of the FLCN gene identify six novel variants and a Danish founder mutation.J Hum Genet. 2017 Feb;62(2):151-157. doi: 10.1038/jhg.2016.118. Epub 2016 Oct 13. J Hum Genet. 2017. PMID: 27734835
-
Adding In Silico Assessment of Potential Splice Aberration to the Integrated Evaluation of BRCA Gene Unclassified Variants.Hum Mutat. 2016 Jul;37(7):627-39. doi: 10.1002/humu.22973. Epub 2016 Apr 15. Hum Mutat. 2016. PMID: 26913838 Free PMC article.
-
Identification of eight novel SDHB, SDHC, SDHD germline variants in Danish pheochromocytoma/paraganglioma patients.Hered Cancer Clin Pract. 2016 Jun 8;14:13. doi: 10.1186/s13053-016-0053-6. eCollection 2016. Hered Cancer Clin Pract. 2016. PMID: 27279923 Free PMC article.
-
Genome-wide analysis of alternative transcripts in human breast cancer.Breast Cancer Res Treat. 2015 Jun;151(2):295-307. doi: 10.1007/s10549-015-3395-2. Epub 2015 Apr 26. Breast Cancer Res Treat. 2015. PMID: 25913416 Free PMC article.
References
-
- Spurdle AB, Healey S, Devereau A, et al. ENIGMA–evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33(1):2–7. doi: 10.1002/humu.21628. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

