Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr Virus lytic activation in B lymphocytes
- PMID: 25717101
- PMCID: PMC4403452
- DOI: 10.1128/JVI.00121-15
Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr Virus lytic activation in B lymphocytes
Abstract
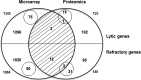
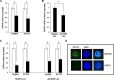
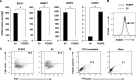
A major hurdle to killing Epstein-Barr virus (EBV)-infected tumor cells using oncolytic therapy is the presence of a substantial fraction of EBV-infected cells that does not support the lytic phase of EBV despite exposure to lytic cycle-promoting agents. To determine the mechanism(s) underlying this refractory state, we developed a strategy to separate lytic from refractory EBV-positive (EBV(+)) cells. By examining the cellular transcriptome in separated cells, we previously discovered that high levels of host STAT3 (signal transducer and activator of transcription 3) curtail the susceptibility of latently infected cells to lytic cycle activation signals. The goals of the present study were 2-fold: (i) to determine the mechanism of STAT3-mediated resistance to lytic activation and (ii) to exploit our findings to enhance susceptibility to lytic activation. We therefore analyzed our microarray data set, cellular proteomes of separated lytic and refractory cells, and a publically available STAT3 chromatin immunoprecipitation sequencing (ChIP-Seq) data set to identify cellular PCBP2 [poly(C)-binding protein 2], an RNA-binding protein, as a transcriptional target of STAT3 in refractory cells. Using Burkitt lymphoma cells and EBV(+) cell lines from patients with hypomorphic STAT3 mutations, we demonstrate that single cells expressing high levels of PCBP2 are refractory to spontaneous and induced EBV lytic activation, STAT3 functions via cellular PCBP2 to regulate lytic susceptibility, and suppression of PCBP2 levels is sufficient to increase the number of EBV lytic cells. We expect that these findings and the genome-wide resources that they provide will accelerate our understanding of a longstanding mystery in EBV biology and guide efforts to improve oncolytic therapy for EBV-associated cancers.
Importance: Most humans are infected with Epstein-Barr virus (EBV), a cancer-causing virus. While EBV generally persists silently in B lymphocytes, periodic lytic (re)activation of latent virus is central to its life cycle and to most EBV-related diseases. However, a substantial fraction of EBV-infected B cells and tumor cells in a population is refractory to lytic activation. This resistance to lytic activation directly and profoundly impacts viral persistence and the effectiveness of oncolytic therapy for EBV(+) cancers. To identify the mechanisms that underlie susceptibility to EBV lytic activation, we used host gene and protein expression profiling of separated lytic and refractory cells. We find that STAT3, a transcription factor overactive in many cancers, regulates PCBP2, a protein important in RNA biogenesis, to regulate susceptibility to lytic cycle activation signals. These findings advance our understanding of EBV persistence and provide important leads on devising methods to improve viral oncolytic therapies.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
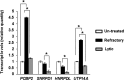


Similar articles
-
STAT3 Regulates Lytic Activation of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2015 Nov;89(22):11347-55. doi: 10.1128/JVI.02008-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339061 Free PMC article.
-
Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes.J Virol. 2013 Nov;87(21):11438-46. doi: 10.1128/JVI.01762-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966384 Free PMC article.
-
Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors.J Virol. 2010 Jan;84(2):993-1004. doi: 10.1128/JVI.01745-09. Epub 2009 Nov 4. J Virol. 2010. PMID: 19889776 Free PMC article.
-
Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review.
-
Human B cells on their route to latent infection--early but transient expression of lytic genes of Epstein-Barr virus.Eur J Cell Biol. 2012 Jan;91(1):65-9. doi: 10.1016/j.ejcb.2011.01.014. Epub 2011 Mar 29. Eur J Cell Biol. 2012. PMID: 21450364 Review.
Cited by
-
KRAB-ZFP Repressors Enforce Quiescence of Oncogenic Human Herpesviruses.J Virol. 2018 Jun 29;92(14):e00298-18. doi: 10.1128/JVI.00298-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29695433 Free PMC article.
-
Epstein-Barr Virus (EBV) Tegument Protein BGLF2 Suppresses Type I Interferon Signaling To Promote EBV Reactivation.J Virol. 2020 May 18;94(11):e00258-20. doi: 10.1128/JVI.00258-20. Print 2020 May 18. J Virol. 2020. PMID: 32213613 Free PMC article.
-
Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction.PLoS Pathog. 2018 Jan 22;14(1):e1006868. doi: 10.1371/journal.ppat.1006868. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29357389 Free PMC article.
-
Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK-STAT pathway.Blood Cancer Discov. 2020 Jul;1(1):112-125. doi: 10.1158/2643-3230.BCD-20-0051. Blood Cancer Discov. 2020. PMID: 33225311 Free PMC article.
-
The Dynamic Interface of Viruses with STATs.J Virol. 2020 Oct 27;94(22):e00856-20. doi: 10.1128/JVI.00856-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847860 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

