Using common spatial distributions of atoms to relate functionally divergent influenza virus N10 and N11 protein structures to functionally characterized neuraminidase structures, toxin cell entry domains, and non-influenza virus cell entry domains
- PMID: 25706124
- PMCID: PMC4337911
- DOI: 10.1371/journal.pone.0117499
Using common spatial distributions of atoms to relate functionally divergent influenza virus N10 and N11 protein structures to functionally characterized neuraminidase structures, toxin cell entry domains, and non-influenza virus cell entry domains
Abstract
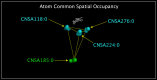
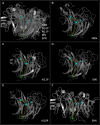
The ability to identify the functional correlates of structural and sequence variation in proteins is a critical capability. We related structures of influenza A N10 and N11 proteins that have no established function to structures of proteins with known function by identifying spatially conserved atoms. We identified atoms with common distributed spatial occupancy in PDB structures of N10 protein, N11 protein, an influenza A neuraminidase, an influenza B neuraminidase, and a bacterial neuraminidase. By superposing these spatially conserved atoms, we aligned the structures and associated molecules. We report spatially and sequence invariant residues in the aligned structures. Spatially invariant residues in the N6 and influenza B neuraminidase active sites were found in previously unidentified spatially equivalent sites in the N10 and N11 proteins. We found the corresponding secondary and tertiary structures of the aligned proteins to be largely identical despite significant sequence divergence. We found structural precedent in known non-neuraminidase structures for residues exhibiting structural and sequence divergence in the aligned structures. In N10 protein, we identified staphylococcal enterotoxin I-like domains. In N11 protein, we identified hepatitis E E2S-like domains, SARS spike protein-like domains, and toxin components shared by alpha-bungarotoxin, staphylococcal enterotoxin I, anthrax lethal factor, clostridium botulinum neurotoxin, and clostridium tetanus toxin. The presence of active site components common to the N6, influenza B, and S. pneumoniae neuraminidases in the N10 and N11 proteins, combined with the absence of apparent neuraminidase function, suggests that the role of neuraminidases in H17N10 and H18N11 emerging influenza A viruses may have changed. The presentation of E2S-like, SARS spike protein-like, or toxin-like domains by the N10 and N11 proteins in these emerging viruses may indicate that H17N10 and H18N11 sialidase-facilitated cell entry has been supplemented or replaced by sialidase-independent receptor binding to an expanded cell population that may include neurons and T-cells.
Conflict of interest statement
Figures






Similar articles
-
Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18903-8. doi: 10.1073/pnas.1212579109. Epub 2012 Sep 24. Proc Natl Acad Sci U S A. 2012. PMID: 23012478 Free PMC article.
-
Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18897-902. doi: 10.1073/pnas.1211037109. Epub 2012 Sep 24. Proc Natl Acad Sci U S A. 2012. PMID: 23012237 Free PMC article.
-
Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.J Virol. 2014 Aug;88(16):9197-207. doi: 10.1128/JVI.00805-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899180 Free PMC article.
-
Bat-derived influenza-like viruses H17N10 and H18N11.Trends Microbiol. 2014 Apr;22(4):183-91. doi: 10.1016/j.tim.2014.01.010. Epub 2014 Feb 26. Trends Microbiol. 2014. PMID: 24582528 Free PMC article. Review.
-
Palmitoylation of influenza virus proteins.Biochem Soc Trans. 2013 Feb 1;41(1):50-5. doi: 10.1042/BST20120210. Biochem Soc Trans. 2013. PMID: 23356257 Review.
Cited by
-
Bat-Borne Influenza A Viruses: An Awakening.Cold Spring Harb Perspect Med. 2021 Feb 1;11(2):a038612. doi: 10.1101/cshperspect.a038612. Cold Spring Harb Perspect Med. 2021. PMID: 31871229 Free PMC article. Review.
References
-
- Chan MCW, Chan RWY, Chan LLY, Mok CKP, Hui KPY, et al. (2013) Tropism and innate host responses of a novel influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory track. Lancet Respir. Med. 1(7):534–542. Available: 10.1016/S2213-2600(13)70138-3 10.1016/S2213-2600(13)70138-3 - DOI - DOI - PMC - PubMed
-
- Wibawa H, Bingham J, Nuradji H, Lowther S, Payne J, et al. (2014) Experimentally Infected Domestic Ducks Show Efficient Transmission of Indonesian H5N1 Highly Pathogenic Avian Influenza Virus, but Lack Persistent Viral Shedding. PloS One 9(1):e83417 Available: 10.1371/journal.pone.0083417 10.1371/journal.pone.0083417 - DOI - DOI - PMC - PubMed
-
- Schrauwen EJA, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM, et al. (2013) Reassortment between Avian H5N1 and Human Influenza Viruses Is Mainly Restricted to the Matrix and Neuraminidase Gene Segments. PloS One 8(3):e59889 Available: 10.1371/journal.pone.0059889 10.1371/journal.pone.0059889 - DOI - DOI - PMC - PubMed
-
- Protein Data Bank ID: 4FVK. Available: http://www.rcsb.org/pdb/explore/explore.do?structureId=4FVK Li Q, Sun XM, Li ZX, Liu Y, Vavricka CJ, et al. (2012) Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc. Natl. Acad. Sci. USA. 109:18897–18902. Available: 10.1073/pnas.1211037109 10.1073/pnas.1211037109 - DOI - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

