Role of epigenetic modifications in luminal breast cancer
- PMID: 25689414
- PMCID: PMC4539290
- DOI: 10.2217/epi.15.10
Role of epigenetic modifications in luminal breast cancer
Abstract
Luminal breast cancers represent approximately 75% of cases. Explanations into the causes of endocrine resistance are complex and are generally ascribed to genomic mechanisms. Recently, attention has been drawn to the role of epigenetic modifications in hormone resistance. We review these here. Epigenetic modifications are reversible, heritable and include changes in DNA methylation patterns, modification of histones and altered microRNA expression levels that target the receptors or their signaling pathways. Large-scale analyses indicate distinct epigenomic profiles that distinguish breast cancers from normal and benign tissues. Taking advantage of the reversibility of epigenetic modifications, drugs that target epigenetic modifiers, given in combination with chemotherapies or endocrine therapies, may represent promising approaches to restoration of therapy responsiveness in these cases.
Keywords: breast cancer; epigenetic modifications; estrogen receptors; progesterone receptors.
Conflict of interest statement
The authors are grateful for support from the NIH (RO1-CA026869-35), the Breast Cancer Research Foundation and the National Foundation for Cancer Research. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures
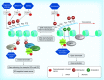
Similar articles
-
Current and upcoming approaches to exploit the reversibility of epigenetic mutations in breast cancer.Breast Cancer Res. 2014 Jul 29;16(4):412. doi: 10.1186/s13058-014-0412-z. Breast Cancer Res. 2014. PMID: 25410383 Free PMC article. Review.
-
Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model.Clin Epigenetics. 2016 Mar 31;8:34. doi: 10.1186/s13148-016-0201-x. eCollection 2016. Clin Epigenetics. 2016. PMID: 27034728 Free PMC article.
-
Epigenetic mechanisms of breast cancer: an update of the current knowledge.Epigenomics. 2014;6(6):651-64. doi: 10.2217/epi.14.59. Epigenomics. 2014. PMID: 25531258 Review.
-
Comparative epigenetic analyses reveal distinct patterns of oncogenic pathways activation in breast cancer subtypes.Hum Mol Genet. 2014 Oct 15;23(20):5378-93. doi: 10.1093/hmg/ddu256. Epub 2014 May 27. Hum Mol Genet. 2014. PMID: 24871326
-
Epigenetic alterations in cancer.Front Biosci (Landmark Ed). 2020 Mar 1;25(6):1058-1109. doi: 10.2741/4847. Front Biosci (Landmark Ed). 2020. PMID: 32114424 Review.
Cited by
-
Effect of Withaferin-A, Withanone, and Caffeic Acid Phenethyl Ester on DNA Methyltransferases: Potential in Epigenetic Cancer Therapy.Curr Top Med Chem. 2024;24(4):379-391. doi: 10.2174/1568026623666230726105017. Curr Top Med Chem. 2024. PMID: 37496252
-
Pan-cancer analysis of m5C regulator genes reveals consistent epigenetic landscape changes in multiple cancers.World J Surg Oncol. 2021 Jul 29;19(1):224. doi: 10.1186/s12957-021-02342-y. World J Surg Oncol. 2021. PMID: 34325709 Free PMC article.
-
Epigenetics in Inflammatory Breast Cancer: Biological Features and Therapeutic Perspectives.Cells. 2020 May 8;9(5):1164. doi: 10.3390/cells9051164. Cells. 2020. PMID: 32397183 Free PMC article. Review.
-
Response to Yang et. al.J Mol Med (Berl). 2019 May;97(5):739-740. doi: 10.1007/s00109-019-01775-z. Epub 2019 Mar 23. J Mol Med (Berl). 2019. PMID: 30903230 No abstract available.
-
Akt regulates RSK2 to alter phosphorylation level of H2A.X in breast cancer.Oncol Lett. 2021 Mar;21(3):187. doi: 10.3892/ol.2021.12448. Epub 2021 Jan 6. Oncol Lett. 2021. PMID: 33574926 Free PMC article.
References
-
- Prat A, Karginova O, Parker JS, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat. 2013;142(2):237–255. - PMC - PubMed
-
•• Data presented in this paper help to improve our understanding of the wide range of breast cancer cell line models through the appropriate pairing of cell lines with relevant
in vivo tumor and normal cell counterpart.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
