Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins
- PMID: 25680966
- PMCID: PMC4357707
- DOI: 10.1093/nar/gkv102
Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins
Abstract
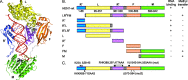
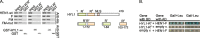
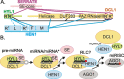
Methylation of 3'-terminal nucleotides of miRNA/miRNA* is part of miRNAs biogenesis in plants but is not found in animals. In Arabidopsis thaliana this reaction is carried out by a multidomain AdoMet-dependent 2'-O-methyltransferase HEN1. Using deletion and structure-guided mutational analysis, we show that the double-stranded RNA-binding domains R(1) and R(2) of HEN1 make significant but uneven contributions to substrate RNA binding, and map residues in each domain responsible for this function. Using GST pull-down assays and yeast two-hybrid analysis we demonstrate direct HEN1 interactions, mediated by its FK506-binding protein-like domain and R(2) domain, with the microRNA biogenesis protein HYL1. Furthermore, we find that HEN1 forms a complex with DICER-LIKE 1 (DCL1) ribonuclease, another key protein involved in miRNA biogenesis machinery. In contrast, no direct interaction is detectable between HEN1 and SERRATE. On the basis of these findings, we propose a mechanism of plant miRNA maturation which involves binding of the HEN1 methyltransferase to the DCL1•HYL1•miRNA complex excluding the SERRATE protein.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
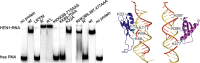




Similar articles
-
SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex.PLoS Genet. 2008 Jun 13;4(6):e1000096. doi: 10.1371/journal.pgen.1000096. PLoS Genet. 2008. PMID: 18551175 Free PMC article.
-
A dominant mutation in DCL1 suppresses the hyl1 mutant phenotype by promoting the processing of miRNA.RNA. 2009 Mar;15(3):450-8. doi: 10.1261/rna.1297109. Epub 2009 Jan 20. RNA. 2009. PMID: 19155326 Free PMC article.
-
Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis.BMC Evol Biol. 2006 Jan 24;6:6. doi: 10.1186/1471-2148-6-6. BMC Evol Biol. 2006. PMID: 16433904 Free PMC article.
-
MicroRNA biogenesis and function in plants.FEBS Lett. 2005 Oct 31;579(26):5923-31. doi: 10.1016/j.febslet.2005.07.071. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16144699 Free PMC article. Review.
-
[Plant micro RNA biogenesis].Postepy Biochem. 2008;54(3):308-16. Postepy Biochem. 2008. PMID: 19112830 Review. Polish.
Cited by
-
microRNA biogenesis and stabilization in plants.Fundam Res. 2023 Mar 17;3(5):707-717. doi: 10.1016/j.fmre.2023.02.023. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933298 Free PMC article. Review.
-
Recent Insights into Plant miRNA Biogenesis: Multiple Layers of miRNA Level Regulation.Plants (Basel). 2023 Jan 11;12(2):342. doi: 10.3390/plants12020342. Plants (Basel). 2023. PMID: 36679055 Free PMC article. Review.
-
Biogenesis, conservation, and function of miRNA in liverworts.J Exp Bot. 2022 Jul 16;73(13):4528-4545. doi: 10.1093/jxb/erac098. J Exp Bot. 2022. PMID: 35275209 Free PMC article. Review.
-
Phylogenetic analyses of seven protein families refine the evolution of small RNA pathways in green plants.Plant Physiol. 2023 May 31;192(2):1183-1203. doi: 10.1093/plphys/kiad141. Plant Physiol. 2023. PMID: 36869858 Free PMC article.
-
Selective chemical tracking of Dnmt1 catalytic activity in live cells.Mol Cell. 2022 Mar 3;82(5):1053-1065.e8. doi: 10.1016/j.molcel.2022.02.008. Mol Cell. 2022. PMID: 35245449 Free PMC article.
References
-
- Bologna N.G., Schapire A.L., Palatnik J.F. Processing of plant microRNA precursors. Brief. Funct. Genomics. 2013;12:37–45. - PubMed
-
- Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005;57:173–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

