Deletion of AU-rich elements within the Bcl2 3'UTR reduces protein expression and B cell survival in vivo
- PMID: 25680182
- PMCID: PMC4332480
- DOI: 10.1371/journal.pone.0116899
Deletion of AU-rich elements within the Bcl2 3'UTR reduces protein expression and B cell survival in vivo
Abstract
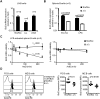
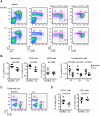
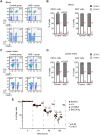
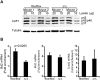
Post-transcriptional mRNA regulation by RNA binding proteins (RBPs) associated with AU-rich elements (AREs) present in the 3' untranslated region (3'UTR) of specific mRNAs modulates transcript stability and translation in eukaryotic cells. Here we have functionally characterised the importance of the AREs present within the Bcl2 3'UTR in order to maintain Bcl2 expression. Gene targeting deletion of 300 nucleotides of the Bcl2 3'UTR rich in AREs diminishes Bcl2 mRNA stability and protein levels in primary B cells, decreasing cell lifespan. Generation of chimeric mice indicates that Bcl2-ARE∆/∆ B cells have an intrinsic competitive disadvantage compared to wild type cells. Biochemical assays and predictions using a bioinformatics approach show that several RBPs bind to the Bcl2 AREs, including AUF1 and HuR proteins. Altogether, association of RBPs to Bcl2 AREs contributes to Bcl2 protein expression by stabilizing Bcl2 mRNA and promotes B cell maintenance.
Conflict of interest statement
Figures



Similar articles
-
Interaction between 3' untranslated region of calcitonin receptor messenger ribonucleic acid (RNA) and adenylate/uridylate (AU)-rich element binding proteins (AU-rich RNA-binding factor 1 and Hu antigen R).Endocrinology. 2004 Apr;145(4):1730-8. doi: 10.1210/en.2003-0862. Epub 2004 Jan 8. Endocrinology. 2004. PMID: 14715706
-
Regulation of thrombospondin-1 expression through AU-rich elements in the 3'UTR of the mRNA.Cell Mol Biol Lett. 2011 Mar;16(1):55-68. doi: 10.2478/s11658-010-0037-x. Epub 2010 Dec 15. Cell Mol Biol Lett. 2011. PMID: 21161418 Free PMC article.
-
Autotaxin Expression Is Regulated at the Post-transcriptional Level by the RNA-binding Proteins HuR and AUF1.J Biol Chem. 2016 Dec 9;291(50):25823-25836. doi: 10.1074/jbc.M116.756908. Epub 2016 Oct 26. J Biol Chem. 2016. PMID: 27784781 Free PMC article.
-
Physiological networks and disease functions of RNA-binding protein AUF1.Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):549-64. doi: 10.1002/wrna.1230. Epub 2014 Mar 28. Wiley Interdiscip Rev RNA. 2014. PMID: 24687816 Review.
-
Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):680-8. doi: 10.1016/j.bbagrm.2012.12.002. Epub 2012 Dec 14. Biochim Biophys Acta. 2013. PMID: 23246978 Free PMC article. Review.
Cited by
-
Uncovering the Role of RNA-Binding Proteins in Gene Expression in the Immune System.Front Immunol. 2018 May 23;9:1094. doi: 10.3389/fimmu.2018.01094. eCollection 2018. Front Immunol. 2018. PMID: 29875770 Free PMC article. Review.
-
hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma.EMBO J. 2019 Feb 1;38(3):e99128. doi: 10.15252/embj.201899128. Epub 2018 Dec 7. EMBO J. 2019. PMID: 30530525 Free PMC article.
-
Plant 3' Regulatory Regions From mRNA-Encoding Genes and Their Uses to Modulate Expression.Front Plant Sci. 2020 Aug 14;11:1252. doi: 10.3389/fpls.2020.01252. eCollection 2020. Front Plant Sci. 2020. PMID: 32922424 Free PMC article. Review.
-
Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells.Nat Commun. 2017 Sep 13;8(1):530. doi: 10.1038/s41467-017-00454-2. Nat Commun. 2017. PMID: 28904350 Free PMC article.
-
Negative Regulation of BOK Expression by Recruitment of TRIM28 to Regulatory Elements in Its 3' Untranslated Region.iScience. 2018 Nov 30;9:461-474. doi: 10.1016/j.isci.2018.11.005. Epub 2018 Nov 10. iScience. 2018. PMID: 30471638 Free PMC article.
References
-
- Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM (1984) Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 226: 1097–1099. - PubMed
-
- McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, et al. (1989) bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell 57: 79–88. - PubMed
-
- Letai A, Sorcinelli MD, Beard C, Korsmeyer SJ (2004) Antiapoptotic BCL-2 is required for maintenance of a model leukemia. Cancer Cell 6: 241–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

