Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts
- PMID: 25670856
- PMCID: PMC4423675
- DOI: 10.1074/jbc.M114.620641
Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts
Abstract
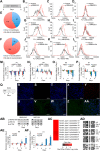
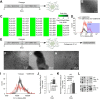
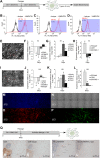
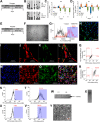
Human embryonic stem cells (hESCs) have been routinely treated with bone morphogenetic protein and/or inhibitors of activin/nodal signaling to obtain cells that express trophoblast markers. Trophoblasts can terminally differentiate to either extravillous trophoblasts or syncytiotrophoblasts. The signaling pathways that govern the terminal fate of these trophoblasts are not understood. We show that activin/nodal signaling switches the terminal fate of these hESC-derived trophoblasts. Inhibition of activin/nodal signaling leads to formation of extravillous trophoblast, whereas loss of activin/nodal inhibition leads to the formation of syncytiotrophoblasts. Also, the ability of hESCs to form bona fide trophoblasts has been intensely debated. We have examined hESC-derived trophoblasts in the light of stringent criteria that were proposed recently, such as hypomethylation of the ELF5-2b promoter region and down-regulation of HLA class I antigens. We report that trophoblasts that possess these properties can indeed be obtained from hESCs.
Keywords: Activin; Activin/Nodal Signaling; BMP Signaling; Bone Morphogenetic Protein (BMP); Differentiation; ELF5; Embryonic Stem Cell; Human Embryonic Stem Cells; Trophoblast.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Comparison of Four Protocols for In Vitro Differentiation of Human Embryonic Stem Cells into Trophoblast Lineages by BMP4 and Dual Inhibition of Activin/Nodal and FGF2 Signaling.Reprod Sci. 2024 Jan;31(1):173-189. doi: 10.1007/s43032-023-01334-5. Epub 2023 Sep 1. Reprod Sci. 2024. PMID: 37658178 Free PMC article.
-
Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells.J Biol Chem. 2008 Sep 5;283(36):24991-5002. doi: 10.1074/jbc.M803893200. Epub 2008 Jul 2. J Biol Chem. 2008. PMID: 18596037 Free PMC article.
-
FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast.Stem Cells Dev. 2012 Nov 1;21(16):2987-3000. doi: 10.1089/scd.2012.0099. Epub 2012 Aug 6. Stem Cells Dev. 2012. PMID: 22724507 Free PMC article.
-
Trophoblast differentiation of human embryonic stem cells.Biotechnol J. 2013 Apr;8(4):421-33. doi: 10.1002/biot.201200203. Epub 2013 Jan 17. Biotechnol J. 2013. PMID: 23325630 Review.
-
The role of activin/nodal and Wnt signaling in endoderm formation.Vitam Horm. 2011;85:207-16. doi: 10.1016/B978-0-12-385961-7.00010-X. Vitam Horm. 2011. PMID: 21353882 Review.
Cited by
-
Comparison of Four Protocols for In Vitro Differentiation of Human Embryonic Stem Cells into Trophoblast Lineages by BMP4 and Dual Inhibition of Activin/Nodal and FGF2 Signaling.Reprod Sci. 2024 Jan;31(1):173-189. doi: 10.1007/s43032-023-01334-5. Epub 2023 Sep 1. Reprod Sci. 2024. PMID: 37658178 Free PMC article.
-
Modeling human trophoblast, the placental epithelium at the maternal fetal interface.Reproduction. 2020 Jul;160(1):R1-R11. doi: 10.1530/REP-19-0428. Reproduction. 2020. PMID: 32485667 Free PMC article. Review.
-
Sex-Specific Gene Expression Differences Are Evident in Human Embryonic Stem Cells and During In Vitro Differentiation of Human Placental Progenitor Cells.Stem Cells Dev. 2018 Oct 1;27(19):1360-1375. doi: 10.1089/scd.2018.0081. Epub 2018 Aug 21. Stem Cells Dev. 2018. PMID: 29993333 Free PMC article.
-
Trophoblast Differentiation: Mechanisms and Implications for Pregnancy Complications.Nutrients. 2023 Aug 12;15(16):3564. doi: 10.3390/nu15163564. Nutrients. 2023. PMID: 37630754 Free PMC article. Review.
-
Omics Approaches to Study Formation and Function of Human Placental Syncytiotrophoblast.Front Cell Dev Biol. 2021 Jun 15;9:674162. doi: 10.3389/fcell.2021.674162. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34211975 Free PMC article. Review.
References
-
- Xu R.-H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 - PubMed
-
- Gerami-Naini B., Dovzhenko O. V., Durning M., Wegner F. H., Thomson J. A., Golos T. G. (2004) Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology 145, 1517–1524 - PubMed
-
- Matin M. M., Walsh J. R., Gokhale P. J., Draper J. S., Bahrami A. R., Morton I., Moore H. D., Andrews P. W. (2004) Specific knockdown of Oct4 and β2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells 22, 659–668 - PubMed
-
- Hay D. C., Sutherland L., Clark J., Burdon T. (2004) Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 22, 225–235 - PubMed
-
- Hyslop L., Stojkovic M., Armstrong L., Walter T., Stojkovic P., Przyborski S., Herbert M., Murdoch A., Strachan T., Lako M. (2005) Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 23, 1035–1043 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

