Evolution of p53 transactivation specificity through the lens of a yeast-based functional assay
- PMID: 25668429
- PMCID: PMC4323202
- DOI: 10.1371/journal.pone.0116177
Evolution of p53 transactivation specificity through the lens of a yeast-based functional assay
Abstract
Co-evolution of transcription factors (TFs) with their respective cis-regulatory network enhances functional diversity in the course of evolution. We present a new approach to investigate transactivation capacity of sequence-specific TFs in evolutionary studies. Saccharomyces cerevisiae was used as an in vivo test tube and p53 proteins derived from human and five commonly used animal models were chosen as proof of concept. p53 is a highly conserved master regulator of environmental stress responses. Previous reports indicated conserved p53 DNA binding specificity in vitro, even for evolutionary distant species. We used isogenic yeast strains where p53-dependent transactivation was measured towards chromosomally integrated p53 response elements (REs). Ten REs were chosen to sample a wide range of DNA binding affinity and transactivation capacity for human p53 and proteins were expressed at two levels using an inducible expression system. We showed that the assay is amenable to study thermo-sensitivity of frog p53, and that chimeric constructs containing an ectopic transactivation domain could be rapidly developed to enhance the activity of proteins, such as fruit fly p53, that are poorly effective in engaging the yeast transcriptional machinery. Changes in the profile of relative transactivation towards the ten REs were measured for each p53 protein and compared to the profile obtained with human p53. These results, which are largely independent from relative p53 protein levels, revealed widespread evolutionary divergence of p53 transactivation specificity, even between human and mouse p53. Fruit fly and human p53 exhibited the largest discrimination among REs while zebrafish p53 was the least selective.
Conflict of interest statement
Figures

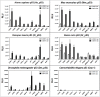
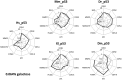
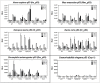

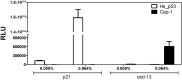
Similar articles
-
Functional evolution of the p53 regulatory network through its target response elements.Proc Natl Acad Sci U S A. 2008 Jan 22;105(3):944-9. doi: 10.1073/pnas.0704694105. Epub 2008 Jan 10. Proc Natl Acad Sci U S A. 2008. PMID: 18187580 Free PMC article.
-
Noncanonical DNA motifs as transactivation targets by wild type and mutant p53.PLoS Genet. 2008 Jun 27;4(6):e1000104. doi: 10.1371/journal.pgen.1000104. PLoS Genet. 2008. PMID: 18714371 Free PMC article.
-
Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants.Oncogene. 2001 Jun 7;20(26):3409-19. doi: 10.1038/sj.onc.1204457. Oncogene. 2001. PMID: 11423991
-
Potentiating the p53 network.Discov Med. 2010 Jul;10(50):94-100. Discov Med. 2010. PMID: 20670604 Review.
-
Human transcription factors in yeast: the fruitful examples of P53 and NF-кB.FEMS Yeast Res. 2016 Nov;16(7):fow083. doi: 10.1093/femsyr/fow083. Epub 2016 Sep 27. FEMS Yeast Res. 2016. PMID: 27683095 Review.
Cited by
-
Retrotransposon-derived p53 binding sites enhance telomere maintenance and genome protection.Bioessays. 2016 Oct;38(10):943-9. doi: 10.1002/bies.201600078. Epub 2016 Aug 19. Bioessays. 2016. PMID: 27539745 Free PMC article. Review.
-
Tumor suppressor p53: from engaging DNA to target gene regulation.Nucleic Acids Res. 2020 Sep 18;48(16):8848-8869. doi: 10.1093/nar/gkaa666. Nucleic Acids Res. 2020. PMID: 32797160 Free PMC article.
-
p73, like its p53 homolog, shows preference for inverted repeats forming cruciforms.PLoS One. 2018 Apr 18;13(4):e0195835. doi: 10.1371/journal.pone.0195835. eCollection 2018. PLoS One. 2018. PMID: 29668749 Free PMC article.
-
p53, p63 and p73 in the wonderland of S. cerevisiae.Oncotarget. 2017 Jun 16;8(34):57855-57869. doi: 10.18632/oncotarget.18506. eCollection 2017 Aug 22. Oncotarget. 2017. PMID: 28915717 Free PMC article. Review.
-
Extracellular and Intracellular Factors in Brain Cancer.Front Cell Dev Biol. 2021 Aug 27;9:699103. doi: 10.3389/fcell.2021.699103. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513834 Free PMC article. Review.
References
-
- Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4: 793–805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

