The multivesicular body is the major internal site of prion conversion
- PMID: 25663703
- PMCID: PMC4379730
- DOI: 10.1242/jcs.165472
The multivesicular body is the major internal site of prion conversion
Abstract
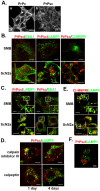
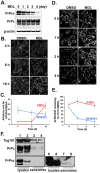
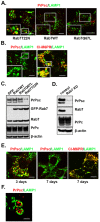
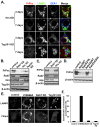
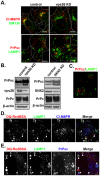
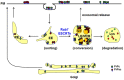
The conversion of the properly folded prion protein, PrPc, to its misfolded amyloid form, PrPsc, occurs as the two proteins traffic along the endocytic pathway and PrPc is exposed to PrPsc. To determine the specific site of prion conversion, we knocked down various proteins in the endocytic pathway including Rab7a, Tsg101 and Hrs (also known as HGS). PrPsc was markedly reduced in two chronically infected cell lines by preventing the maturation of the multivesicular body, a process that begins in the early endosome and ends with the sorting of cargo to the lysosome. By contrast, knocking down proteins in the retromer complex, which diverts cargo away from the multivesicular body caused an increase in PrPsc levels. These results suggest that the multivesicular body is the major site for intracellular conversion of PrPc to PrPsc.
Keywords: Conversion; Multivesicular body; Prion; Scrapie.
© 2015. Published by The Company of Biologists Ltd.
Figures
Similar articles
-
Prions amplify through degradation of the VPS10P sorting receptor sortilin.PLoS Pathog. 2017 Jun 30;13(6):e1006470. doi: 10.1371/journal.ppat.1006470. eCollection 2017 Jun. PLoS Pathog. 2017. PMID: 28665987 Free PMC article.
-
Sheep scrapie susceptibility-linked polymorphisms do not modulate the initial binding of cellular to disease-associated prion protein prior to conversion.J Gen Virol. 2005 Sep;86(Pt 9):2627-2634. doi: 10.1099/vir.0.80901-0. J Gen Virol. 2005. PMID: 16099922
-
Docosahexaenoic and eicosapentaenoic acids increase prion formation in neuronal cells.BMC Biol. 2008 Sep 12;6:39. doi: 10.1186/1741-7007-6-39. BMC Biol. 2008. PMID: 18789130 Free PMC article.
-
Folding and misfolding of the prion protein in the secretory pathway.Amyloid. 2004 Sep;11(3):162-72. doi: 10.1080/1350-6120400000723. Amyloid. 2004. PMID: 15523918 Review.
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
Cited by
-
Efficient inhibition of infectious prions multiplication and release by targeting the exosomal pathway.Cell Mol Life Sci. 2015 Nov;72(22):4409-27. doi: 10.1007/s00018-015-1945-8. Epub 2015 Jun 6. Cell Mol Life Sci. 2015. PMID: 26047659 Free PMC article.
-
NMDA Receptor and L-Type Calcium Channel Modulate Prion Formation.Cell Mol Neurobiol. 2021 Jan;41(1):191-198. doi: 10.1007/s10571-020-00834-1. Epub 2020 Apr 1. Cell Mol Neurobiol. 2021. PMID: 32239389 Free PMC article.
-
Prion infection impairs lysosomal degradation capacity by interfering with rab7 membrane attachment in neuronal cells.Sci Rep. 2016 Feb 11;6:21658. doi: 10.1038/srep21658. Sci Rep. 2016. PMID: 26865414 Free PMC article.
-
Axonal changes in experimental prion diseases recapitulate those following constriction of postganglionic branches of the superior cervical ganglion: a comparison 40 years later.Prion. 2019 Jan;13(1):83-93. doi: 10.1080/19336896.2019.1595315. Prion. 2019. PMID: 30966865 Free PMC article. Review.
-
Cellular mechanisms responsible for cell-to-cell spreading of prions.Cell Mol Life Sci. 2018 Jul;75(14):2557-2574. doi: 10.1007/s00018-018-2823-y. Epub 2018 May 14. Cell Mol Life Sci. 2018. PMID: 29761205 Free PMC article. Review.
References
-
- Ali N., Zhang L., Taylor S., Mironov A., Urbé S., Woodman P. (2013). Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr. Biol. 23, 453–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

