Flexibility and small pockets at protein-protein interfaces: New insights into druggability
- PMID: 25662442
- PMCID: PMC4726663
- DOI: 10.1016/j.pbiomolbio.2015.01.009
Flexibility and small pockets at protein-protein interfaces: New insights into druggability
Abstract
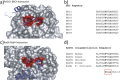
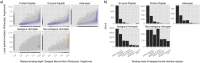
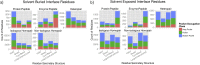
The transient assembly of multiprotein complexes mediates many aspects of cell regulation and signalling in living organisms. Modulation of the formation of these complexes through targeting protein-protein interfaces can offer greater selectivity than the inhibition of protein kinases, proteases or other post-translational regulatory enzymes using substrate, co-factor or transition state mimetics. However, capitalising on protein-protein interaction interfaces as drug targets has been hindered by the nature of interfaces that tend to offer binding sites lacking the well-defined large cavities of classical drug targets. In this review we posit that interfaces formed by concerted folding and binding (disorder-to-order transitions on binding) of one partner and other examples of interfaces where a protein partner is bound through a continuous epitope from a surface-exposed helix, flexible loop or chain extension may be more tractable for the development of "orthosteric", competitive chemical modulators; these interfaces tend to offer small-volume but deep pockets and/or larger grooves that may be bound tightly by small chemical entities. We discuss examples of such protein-protein interaction interfaces for which successful chemical modulators are being developed.
Keywords: Hotspots; Inhibitors druggability; Protein–protein interfaces.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

Similar articles
-
Structural biology and drug discovery for protein-protein interactions.Trends Pharmacol Sci. 2012 May;33(5):241-8. doi: 10.1016/j.tips.2012.03.006. Epub 2012 Apr 11. Trends Pharmacol Sci. 2012. PMID: 22503442 Review.
-
Druggability of dynamic protein-protein interfaces.Curr Pharm Des. 2012;18(30):4599-606. doi: 10.2174/138161212802651652. Curr Pharm Des. 2012. PMID: 22650258 Review.
-
Structural conservation of druggable hot spots in protein-protein interfaces.Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):13528-33. doi: 10.1073/pnas.1101835108. Epub 2011 Aug 1. Proc Natl Acad Sci U S A. 2011. PMID: 21808046 Free PMC article.
-
Structural biology and drug discovery of difficult targets: the limits of ligandability.Chem Biol. 2012 Jan 27;19(1):42-50. doi: 10.1016/j.chembiol.2011.12.013. Chem Biol. 2012. PMID: 22284353 Review.
-
Modulating protein-protein interactions: from structural determinants of binding to druggability prediction to application.Curr Pharm Des. 2012;18(30):4630-47. doi: 10.2174/138161212802651553. Curr Pharm Des. 2012. PMID: 22650257 Review.
Cited by
-
mCSM-PPI2: predicting the effects of mutations on protein-protein interactions.Nucleic Acids Res. 2019 Jul 2;47(W1):W338-W344. doi: 10.1093/nar/gkz383. Nucleic Acids Res. 2019. PMID: 31114883 Free PMC article.
-
ppiGReMLIN: a graph mining based detection of conserved structural arrangements in protein-protein interfaces.BMC Bioinformatics. 2020 Apr 15;21(1):143. doi: 10.1186/s12859-020-3474-1. BMC Bioinformatics. 2020. PMID: 32293241 Free PMC article.
-
Protein structure-function continuum model: Emerging nexuses between specificity, evolution, and structure.Protein Sci. 2024 Apr;33(4):e4968. doi: 10.1002/pro.4968. Protein Sci. 2024. PMID: 38532700 Review.
-
Computational prediction of protein interfaces: A review of data driven methods.FEBS Lett. 2015 Nov 30;589(23):3516-26. doi: 10.1016/j.febslet.2015.10.003. Epub 2015 Oct 13. FEBS Lett. 2015. PMID: 26460190 Free PMC article. Review.
-
Familial STAG2 germline mutation defines a new human cohesinopathy.NPJ Genom Med. 2017 Mar 20;2:7. doi: 10.1038/s41525-017-0009-4. eCollection 2017. NPJ Genom Med. 2017. PMID: 29263825 Free PMC article.
References
-
- Ben-Shimon A., Eisenstein M. Computational mapping of anchoring spots on protein surfaces. J. Mol. Biol. 2010;402:259–277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

