Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T
- PMID: 25660545
- PMCID: PMC4348146
- DOI: 10.1016/j.cub.2015.01.059
Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T
Abstract
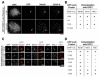
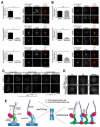
The kinetochore provides a vital connection between chromosomes and spindle microtubules [1, 2]. Defining the molecular architecture of the core kinetochore components is critical for understanding the mechanisms by which the kinetochore directs chromosome segregation. The KNL1/Mis12 complex/Ndc80 complex (KMN) network acts as the primary microtubule-binding interface at kinetochores [3] and provides a platform to recruit regulatory proteins [4]. Recent work found that the inner kinetochore components CENP-C and CENP-T act in parallel to recruit the KMN network to kinetochores [5-8]. However, due to the presence of these dual pathways, it has not been possible to distinguish differences in the nature of kinetochore assembly downstream of CENP-C or CENP-T. Here, we separated these pathways by targeting CENP-C and CENP-T independently to an ectopic chromosomal locus in human cells. Our work reveals that the organization of the KMN network components downstream of CENP-C and CENP-T is distinct. CENP-C recruits the Ndc80 complex through its interactions with KNL1 and the Mis12 complex. In contrast, CENP-T directly interacts with Ndc80, which in turn promotes KNL1/Mis12 complex recruitment through a separate region on CENP-T, resulting in functional relationships for KMN network localization that are inverted relative to the CENP-C pathway. We also find that distinct regulatory paradigms control the assembly of these pathways, with Aurora B kinase promoting KMN network recruitment to CENP-C and cyclin-dependent kinase (CDK) regulating KMN network recruitment to CENP-T. This work reveals unexpected complexity for the architecture and regulation of the core components of the kinetochore-microtubule interface.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
A quantitative description of Ndc80 complex linkage to human kinetochores.Nat Commun. 2015 Sep 8;6:8161. doi: 10.1038/ncomms9161. Nat Commun. 2015. PMID: 26345214 Free PMC article.
-
Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore.Curr Biol. 2011 Mar 8;21(5):391-8. doi: 10.1016/j.cub.2010.12.039. Epub 2011 Feb 25. Curr Biol. 2011. PMID: 21353556 Free PMC article.
-
Multiple phosphorylations control recruitment of the KMN network onto kinetochores.Nat Cell Biol. 2018 Dec;20(12):1378-1388. doi: 10.1038/s41556-018-0230-0. Epub 2018 Nov 12. Nat Cell Biol. 2018. PMID: 30420662
-
Where is the right path heading from the centromere to spindle microtubules?Cell Cycle. 2019 Jun;18(11):1199-1211. doi: 10.1080/15384101.2019.1617008. Epub 2019 May 20. Cell Cycle. 2019. PMID: 31075048 Free PMC article. Review.
-
The ABCs of CENPs.Chromosoma. 2011 Oct;120(5):425-46. doi: 10.1007/s00412-011-0330-0. Epub 2011 Jul 13. Chromosoma. 2011. PMID: 21751032 Review.
Cited by
-
The Four Causes: The Functional Architecture of Centromeres and Kinetochores.Annu Rev Genet. 2022 Nov 30;56:279-314. doi: 10.1146/annurev-genet-072820-034559. Epub 2022 Sep 2. Annu Rev Genet. 2022. PMID: 36055650 Free PMC article. Review.
-
Higher-order protein assembly controls kinetochore formation.Nat Cell Biol. 2024 Jan;26(1):45-56. doi: 10.1038/s41556-023-01313-7. Epub 2024 Jan 2. Nat Cell Biol. 2024. PMID: 38168769 Free PMC article.
-
Physiological relevance of post-translational regulation of the spindle assembly checkpoint protein BubR1.Cell Biosci. 2021 Apr 23;11(1):76. doi: 10.1186/s13578-021-00589-2. Cell Biosci. 2021. PMID: 33892776 Free PMC article. Review.
-
The right place at the right time: Aurora B kinase localization to centromeres and kinetochores.Essays Biochem. 2020 Sep 4;64(2):299-311. doi: 10.1042/EBC20190081. Essays Biochem. 2020. PMID: 32406506 Free PMC article. Review.
-
The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes.Chromosoma. 2017 Jun;126(3):351-364. doi: 10.1007/s00412-016-0618-1. Epub 2016 Nov 11. Chromosoma. 2017. PMID: 27837282 Free PMC article. Review.
References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. - PubMed
-
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

