Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion
- PMID: 25646410
- PMCID: PMC4343099
- DOI: 10.1073/pnas.1414190112
Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion
Abstract
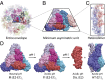
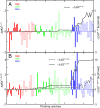
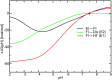
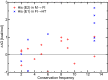
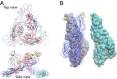
Alphavirus envelope proteins, organized as trimers of E2-E1 heterodimers on the surface of the pathogenic alphavirus, mediate the low pH-triggered fusion of viral and endosomal membranes in human cells. The lack of specific treatment for alphaviral infections motivates our exploration of potential antiviral approaches by inhibiting one or more fusion steps in the common endocytic viral entry pathway. In this work, we performed constant pH molecular dynamics based on an atomic model of the alphavirus envelope with icosahedral symmetry. We have identified pH-sensitive residues that cause the largest shifts in thermodynamic driving forces under neutral and acidic pH conditions for various fusion steps. A series of conserved interdomain His residues is identified to be responsible for the pH-dependent conformational changes in the fusion process, and ligand binding sites in their vicinity are anticipated to be potential drug targets aimed at inhibiting viral infections.
Keywords: alphavirus; constant pH molecular dynamics; envelope protein; membrane fusion; pH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Interactions involved in pH protection of the alphavirus fusion protein.Virology. 2015 Dec;486:173-9. doi: 10.1016/j.virol.2015.08.028. Epub 2015 Oct 2. Virology. 2015. PMID: 26433749 Free PMC article.
-
Acidic pH-Induced Conformational Changes in Chikungunya Virus Fusion Protein E1: a Spring-Twisted Region in the Domain I-III Linker Acts as a Hinge Point for Swiveling Motion of Domains.J Virol. 2020 Nov 9;94(23):e01561-20. doi: 10.1128/JVI.01561-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938768 Free PMC article.
-
Structural changes of envelope proteins during alphavirus fusion.Nature. 2010 Dec 2;468(7324):705-8. doi: 10.1038/nature09546. Nature. 2010. PMID: 21124457 Free PMC article.
-
Entry and uncoating of enveloped viruses.Biochem J. 1994 Sep 1;302 ( Pt 2)(Pt 2):313-20. doi: 10.1042/bj3020313. Biochem J. 1994. PMID: 8092981 Free PMC article. Review. No abstract available.
-
Budding of alphaviruses.Virus Res. 2004 Dec;106(2):103-16. doi: 10.1016/j.virusres.2004.08.008. Virus Res. 2004. PMID: 15567491 Review.
Cited by
-
Cooperative Chikungunya Virus Membrane Fusion and Its Substoichiometric Inhibition by CHK-152 Antibody.Viruses. 2022 Jan 28;14(2):270. doi: 10.3390/v14020270. Viruses. 2022. PMID: 35215863 Free PMC article.
-
Novel Molecular Signatures of Chikungunya Virus in Puerto Rico.P R Health Sci J. 2019 Mar;38(1):27-32. P R Health Sci J. 2019. PMID: 30924912 Free PMC article.
-
All-Atom Molecular Dynamics of Virus Capsids as Drug Targets.J Phys Chem Lett. 2016 May 19;7(10):1836-44. doi: 10.1021/acs.jpclett.6b00517. Epub 2016 May 4. J Phys Chem Lett. 2016. PMID: 27128262 Free PMC article.
-
Atomistic dynamics of a viral infection process: Release of membrane lytic peptides from a non-enveloped virus.Sci Adv. 2021 Apr 14;7(16):eabe1761. doi: 10.1126/sciadv.abe1761. Print 2021 Apr. Sci Adv. 2021. PMID: 33853772 Free PMC article.
-
Scalable Constant pH Molecular Dynamics in GROMACS.J Chem Theory Comput. 2022 Oct 11;18(10):6148-6160. doi: 10.1021/acs.jctc.2c00516. Epub 2022 Sep 21. J Chem Theory Comput. 2022. PMID: 36128977 Free PMC article.
References
-
- Enserink M. Infectious diseases. Chikungunya: No longer a third world disease. Science. 2007;318(5858):1860–1861. - PubMed
-
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8(7):491–500. - PubMed
-
- Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J Gen Virol. 2002;83(Pt 7):1535–1545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

