Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system
- PMID: 25636838
- PMCID: PMC4357945
- DOI: 10.1128/AEM.04023-14
Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system
Erratum in
-
Erratum for Jiang et al., Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System.Appl Environ Microbiol. 2016 May 31;82(12):3693. doi: 10.1128/AEM.01181-16. Print 2016 Jun 15. Appl Environ Microbiol. 2016. PMID: 27247309 Free PMC article. No abstract available.
Abstract
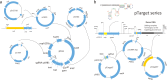
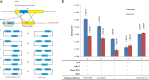
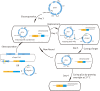
An efficient genome-scale editing tool is required for construction of industrially useful microbes. We describe a targeted, continual multigene editing strategy that was applied to the Escherichia coli genome by using the Streptococcus pyogenes type II CRISPR-Cas9 system to realize a variety of precise genome modifications, including gene deletion and insertion, with a highest efficiency of 100%, which was able to achieve simultaneous multigene editing of up to three targets. The system also demonstrated successful targeted chromosomal deletions in Tatumella citrea, another species of the Enterobacteriaceae, with highest efficiency of 100%.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Efficient Genome Editing in Clostridium cellulolyticum via CRISPR-Cas9 Nickase.Appl Environ Microbiol. 2015 Jul;81(13):4423-31. doi: 10.1128/AEM.00873-15. Epub 2015 Apr 24. Appl Environ Microbiol. 2015. PMID: 25911483 Free PMC article.
-
CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae.FEMS Yeast Res. 2015 Mar;15(2):fov004. doi: 10.1093/femsyr/fov004. Epub 2015 Mar 4. FEMS Yeast Res. 2015. PMID: 25743786 Free PMC article.
-
Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli.Appl Environ Microbiol. 2015 Aug;81(15):5103-14. doi: 10.1128/AEM.01248-15. Epub 2015 May 22. Appl Environ Microbiol. 2015. PMID: 26002895 Free PMC article.
-
Cas9-based tools for targeted genome editing and transcriptional control.Appl Environ Microbiol. 2014 Mar;80(5):1544-52. doi: 10.1128/AEM.03786-13. Epub 2014 Jan 3. Appl Environ Microbiol. 2014. PMID: 24389925 Free PMC article. Review.
-
The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans.J Genet Genomics. 2015 Aug 20;42(8):413-21. doi: 10.1016/j.jgg.2015.06.005. Epub 2015 Jun 26. J Genet Genomics. 2015. PMID: 26336798 Free PMC article. Review.
Cited by
-
Stability of a Mutualistic Escherichia coli Co-Culture During Violacein Production Depends on the Kind of Carbon Source.Eng Life Sci. 2024 Sep 8;24(10):e202400025. doi: 10.1002/elsc.202400025. eCollection 2024 Oct. Eng Life Sci. 2024. PMID: 39391271 Free PMC article.
-
Cytosine Base Editor-Mediated Multiplex Genome Editing to Accelerate Discovery of Novel Antibiotics in Bacillus subtilis and Paenibacillus polymyxa.Front Microbiol. 2021 May 28;12:691839. doi: 10.3389/fmicb.2021.691839. eCollection 2021. Front Microbiol. 2021. PMID: 34122396 Free PMC article.
-
ATP and NADPH engineering of Escherichia coli to improve the production of 4-hydroxyphenylacetic acid using CRISPRi.Biotechnol Biofuels. 2021 Apr 20;14(1):100. doi: 10.1186/s13068-021-01954-6. Biotechnol Biofuels. 2021. PMID: 33879249 Free PMC article.
-
BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site.Nucleic Acids Res. 2019 Jan 10;47(1):253-265. doi: 10.1093/nar/gky1125. Nucleic Acids Res. 2019. PMID: 30418590 Free PMC article.
-
RNase G controls tpiA mRNA abundance in response to oxygen availability in Escherichia coli.J Microbiol. 2019 Oct;57(10):910-917. doi: 10.1007/s12275-019-9354-6. Epub 2019 Sep 30. J Microbiol. 2019. PMID: 31571126
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

