Investigating Hydrophilic Pores in Model Lipid Bilayers Using Molecular Simulations: Correlating Bilayer Properties with Pore-Formation Thermodynamics
- PMID: 25614183
- PMCID: PMC4934177
- DOI: 10.1021/la504049q
Investigating Hydrophilic Pores in Model Lipid Bilayers Using Molecular Simulations: Correlating Bilayer Properties with Pore-Formation Thermodynamics
Abstract
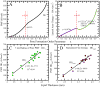
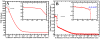
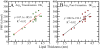
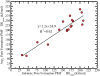
Cell-penetrating and antimicrobial peptides show a remarkable ability to translocate across physiological membranes. Along with factors such as electric-potential-induced perturbations of membrane structure and surface tension effects, experiments invoke porelike membrane configurations during the solute transfer process into vesicles and cells. The initiation and formation of pores are associated with a nontrivial free-energy cost, thus necessitating a consideration of the factors associated with pore formation and the attendant free energies. Because of experimental and modeling challenges related to the long time scales of the translocation process, we use umbrella sampling molecular dynamics simulations with a lipid-density-based order parameter to investigate membrane-pore-formation free energy employing Martini coarse-grained models. We investigate structure and thermodynamic features of the pore in 18 lipids spanning a range of headgroups, charge states, acyl chain lengths, and saturation. We probe the dependence of pore-formation barriers on the area per lipid, lipid bilayer thickness, and membrane bending rigidities in three different lipid classes. The pore-formation free energy in pure bilayers and peptide translocating scenarios are significantly coupled with bilayer thickness. Thicker bilayers require more reversible work to create pores. The pore-formation free energy is higher in peptide-lipid systems than in peptide-free lipid systems due to penalties to maintain the solvation of charged hydrophilic solutes within the membrane environment.
Figures




Similar articles
-
Translocation thermodynamics of linear and cyclic nonaarginine into model DPPC bilayer via coarse-grained molecular dynamics simulation: implications of pore formation and nonadditivity.J Phys Chem B. 2014 Mar 13;118(10):2670-82. doi: 10.1021/jp412600e. Epub 2014 Feb 26. J Phys Chem B. 2014. PMID: 24506488 Free PMC article.
-
The importance of membrane defects-lessons from simulations.Acc Chem Res. 2014 Aug 19;47(8):2244-51. doi: 10.1021/ar4002729. Epub 2014 Jun 3. Acc Chem Res. 2014. PMID: 24892900
-
Thermodynamics of cell-penetrating HIV1 TAT peptide insertion into PC/PS/CHOL model bilayers through transmembrane pores: the roles of cholesterol and anionic lipids.Soft Matter. 2016 Aug 10;12(32):6716-27. doi: 10.1039/c5sm01696g. Soft Matter. 2016. PMID: 27435187
-
Membrane pore formation in atomistic and coarse-grained simulations.Biochim Biophys Acta. 2016 Oct;1858(10):2266-2277. doi: 10.1016/j.bbamem.2015.12.031. Epub 2015 Dec 31. Biochim Biophys Acta. 2016. PMID: 26748016 Review.
-
How Lipid Membranes Affect Pore Forming Toxin Activity.Acc Chem Res. 2015 Dec 15;48(12):3073-9. doi: 10.1021/acs.accounts.5b00403. Epub 2015 Dec 7. Acc Chem Res. 2015. PMID: 26641659 Review.
Cited by
-
Recent Advances of Studies on Cell-Penetrating Peptides Based on Molecular Dynamics Simulations.Cells. 2022 Dec 12;11(24):4016. doi: 10.3390/cells11244016. Cells. 2022. PMID: 36552778 Free PMC article. Review.
-
Mechanistic Understanding from Molecular Dynamics in Pharmaceutical Research 2: Lipid Membrane in Drug Design.Pharmaceuticals (Basel). 2021 Oct 19;14(10):1062. doi: 10.3390/ph14101062. Pharmaceuticals (Basel). 2021. PMID: 34681286 Free PMC article. Review.
-
Influence of Temperature on Molecular Adsorption and Transport at Liposome Surfaces Studied by Molecular Dynamics Simulations and Second Harmonic Generation Spectroscopy.J Phys Chem B. 2021 Sep 23;125(37):10506-10513. doi: 10.1021/acs.jpcb.1c04263. Epub 2021 Sep 8. J Phys Chem B. 2021. PMID: 34495664 Free PMC article.
-
Water Pores in Planar Lipid Bilayers at Fast and Slow Rise of Transmembrane Voltage.Membranes (Basel). 2021 Apr 5;11(4):263. doi: 10.3390/membranes11040263. Membranes (Basel). 2021. PMID: 33916447 Free PMC article.
-
How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane.Molecules. 2019 May 7;24(9):1775. doi: 10.3390/molecules24091775. Molecules. 2019. PMID: 31067828 Free PMC article. Review.
References
-
- Drew Bennett WF, Peter Tieleman D. The Importance of Membrane Defects-Lessons from Simulations. Acc Chem Res. 2014;47:2244–2251. - PubMed
-
- Anderluh G, Lakey JH. Proteins: membrane binding and pore formation. Vol. 677 Springer; 2011. - PubMed
-
- Tabaei SR, Rabe M, Zhdanov VP, Cho NJ, Hook F. Single Vesicle Analysis Reveals Nanoscale Membrane Curvature Selective Pore Formation in Lipid Membranes by an Antiviral-Helical Peptide. Nano Letters. 2012;12:5719–5725. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

