Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia
- PMID: 25612542
- PMCID: PMC4303875
- DOI: 10.1038/srep07989
Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia
Abstract
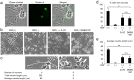
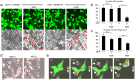
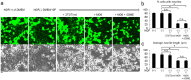
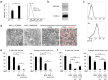
Selective elimination of synaptic connections is a common phenomenon which occurs during both developmental and pathological conditions. Glial cells have a central role in the pruning of synapses by specifically engulfing the degenerating neurites of inappropriate connections, but its regulatory mechanisms have been largely unknown. To identify mediators of this process, we established an in vitro cell culture assay for the synapse elimination. Neuronal differentiation and synapse formation of PC12 cells were induced by culturing the cells with nerve growth factor (NGF) in a serum-free medium. To trigger synapse elimination, the NGF-containing medium was replaced with DMEM containing 10% FBS, and the neurites of PC12 cells degenerated within two days. Co-culturing with MG6 cells, a mouse microglial cell line, accelerated the removal of degenerating neurites of PC12 cells by phagocytosis. When MG6 cells were pre-incubated with exosomes secreted from the differentiated PC12 cells after depolarization, the removal was further accelerated by increasing the expression levels of complement component 3 in the MG6 cells. These results define a role for exosomes as a regulator of synapse elimination and clarify a novel mechanism whereby active synapses promote the pruning of inactive ones by stimulating microglial phagocytosis with exosomes.
Figures

Similar articles
-
Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia.EMBO J. 2020 Aug 17;39(16):e105380. doi: 10.15252/embj.2020105380. Epub 2020 Jul 13. EMBO J. 2020. PMID: 32657463 Free PMC article.
-
Identification of Neuronal Pentraxins as Synaptic Binding Partners of C1q and the Involvement of NP1 in Synaptic Pruning in Adult Mice.Front Immunol. 2021 Feb 8;11:599771. doi: 10.3389/fimmu.2020.599771. eCollection 2020. Front Immunol. 2021. PMID: 33628204 Free PMC article.
-
Complement System in Neural Synapse Elimination in Development and Disease.Adv Immunol. 2017;135:53-79. doi: 10.1016/bs.ai.2017.06.004. Epub 2017 Jul 31. Adv Immunol. 2017. PMID: 28826529 Review.
-
PolyI:C Maternal Immune Activation on E9.5 Causes the Deregulation of Microglia and the Complement System in Mice, Leading to Decreased Synaptic Spine Density.Int J Mol Sci. 2024 May 17;25(10):5480. doi: 10.3390/ijms25105480. Int J Mol Sci. 2024. PMID: 38791517 Free PMC article.
-
Complement and microglia dependent synapse elimination in brain development.WIREs Mech Dis. 2022 May;14(3):e1545. doi: 10.1002/wsbm.1545. Epub 2021 Nov 4. WIREs Mech Dis. 2022. PMID: 34738335 Free PMC article. Review.
Cited by
-
Microglial derived extracellular vesicles activate autophagy and mediate multi-target signaling to maintain cellular homeostasis.J Extracell Vesicles. 2020 Nov;10(1):e12022. doi: 10.1002/jev2.12022. Epub 2020 Nov 25. J Extracell Vesicles. 2020. PMID: 33708355 Free PMC article.
-
Microglial Extracellular Vesicles as Vehicles for Neurodegeneration Spreading.Biomolecules. 2021 May 21;11(6):770. doi: 10.3390/biom11060770. Biomolecules. 2021. PMID: 34063832 Free PMC article. Review.
-
The Role of Extracellular Vesicles in the Developing Brain: Current Perspective and Promising Source of Biomarkers and Therapy for Perinatal Brain Injury.Front Neurosci. 2021 Sep 24;15:744840. doi: 10.3389/fnins.2021.744840. eCollection 2021. Front Neurosci. 2021. PMID: 34630028 Free PMC article. Review.
-
Immunoregulatory Effects of Myeloid-Derived Suppressor Cell Exosomes in Mouse Model of Autoimmune Alopecia Areata.Front Immunol. 2018 Jun 6;9:1279. doi: 10.3389/fimmu.2018.01279. eCollection 2018. Front Immunol. 2018. PMID: 29951053 Free PMC article.
-
Key features of the innate immune response is mediated by the immunoproteasome in microglia.Res Sq [Preprint]. 2024 Jun 6:rs.3.rs-4467983. doi: 10.21203/rs.3.rs-4467983/v1. Res Sq. 2024. PMID: 38883799 Free PMC article. Preprint.
References
-
- Goda Y. & Davis G. W. Mechanisms of synapse assembly and disassembly. Neuron 40, 243–264 (2003). - PubMed
-
- Morris G. P., Clark I. A., Zinn R. & Vissel B. Microglia: a new frontier for synaptic plasticity, learning and memory, and neurodegenerative disease research. Neurobiol Learn Mem 105, 40–53 (2013). - PubMed
-
- Kantor D. B. & Kolodkin A. L. Curbing the excesses of youth: molecular insights into axonal pruning. Neuron 38, 849–852 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

