Plumbagin elicits differential proteomic responses mainly involving cell cycle, apoptosis, autophagy, and epithelial-to-mesenchymal transition pathways in human prostate cancer PC-3 and DU145 cells
- PMID: 25609920
- PMCID: PMC4294653
- DOI: 10.2147/DDDT.S71677
Plumbagin elicits differential proteomic responses mainly involving cell cycle, apoptosis, autophagy, and epithelial-to-mesenchymal transition pathways in human prostate cancer PC-3 and DU145 cells
Abstract
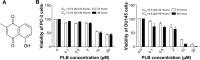
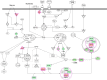
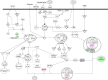
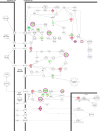
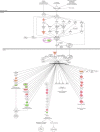
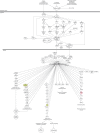
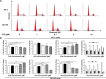
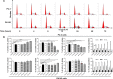
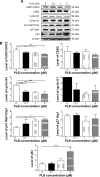
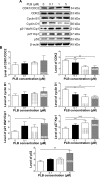
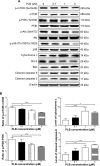
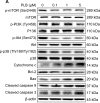
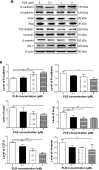
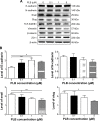
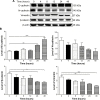
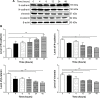
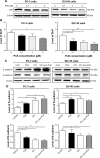
Plumbagin (PLB) has exhibited a potent anticancer effect in preclinical studies, but the molecular interactome remains elusive. This study aimed to compare the quantitative proteomic responses to PLB treatment in human prostate cancer PC-3 and DU145 cells using the approach of stable-isotope labeling by amino acids in cell culture (SILAC). The data were finally validated using Western blot assay. First, the bioinformatic analysis predicted that PLB could interact with 78 proteins that were involved in cell proliferation and apoptosis, immunity, and signal transduction. Our quantitative proteomic study using SILAC revealed that there were at least 1,225 and 267 proteins interacting with PLB and there were 341 and 107 signaling pathways and cellular functions potentially regulated by PLB in PC-3 and DU145 cells, respectively. These proteins and pathways played a critical role in the regulation of cell cycle, apoptosis, autophagy, epithelial to mesenchymal transition (EMT), and reactive oxygen species generation. The proteomic study showed substantial differences in response to PLB treatment between PC-3 and DU145 cells. PLB treatment significantly modulated the expression of critical proteins that regulate cell cycle, apoptosis, and EMT signaling pathways in PC-3 cells but not in DU145 cells. Consistently, our Western blotting analysis validated the bioinformatic and proteomic data and confirmed the modulating effects of PLB on important proteins that regulated cell cycle, apoptosis, autophagy, and EMT in PC-3 and DU145 cells. The data from the Western blot assay could not display significant differences between PC-3 and DU145 cells. These findings indicate that PLB elicits different proteomic responses in PC-3 and DU145 cells involving proteins and pathways that regulate cell cycle, apoptosis, autophagy, reactive oxygen species production, and antioxidation/oxidation homeostasis. This is the first systematic study with integrated computational, proteomic, and functional analyses revealing the networks of signaling pathways and differential proteomic responses to PLB treatment in prostate cancer cells. Quantitative proteomic analysis using SILAC represents an efficient and highly sensitive approach to identify the target networks of anticancer drugs like PLB, and the data may be used to discriminate the molecular and clinical subtypes, and to identify new therapeutic targets and biomarkers, for prostate cancer. Further studies are warranted to explore the potential of quantitative proteomic analysis in the identification of new targets and biomarkers for prostate cancer.
Keywords: EMT; SILAC; proteomics.
Figures








Similar articles
-
Plumbagin induces cell cycle arrest and autophagy and suppresses epithelial to mesenchymal transition involving PI3K/Akt/mTOR-mediated pathway in human pancreatic cancer cells.Drug Des Devel Ther. 2015 Jan 17;9:537-60. doi: 10.2147/DDDT.S73689. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 25632222 Free PMC article.
-
Induction of apoptosis and autophagy via sirtuin1- and PI3K/Akt/mTOR-mediated pathways by plumbagin in human prostate cancer cells.Drug Des Devel Ther. 2015 Mar 12;9:1511-54. doi: 10.2147/DDDT.S75976. eCollection 2015. Drug Des Devel Ther. 2015. Retraction in: Drug Des Devel Ther. 2022 Jul 29;16:2437-2438. doi: 10.2147/DDDT.S383775. PMID: 25834399 Free PMC article. Retracted.
-
Plumbagin suppresses epithelial to mesenchymal transition and stemness via inhibiting Nrf2-mediated signaling pathway in human tongue squamous cell carcinoma cells.Drug Des Devel Ther. 2015 Oct 5;9:5511-51. doi: 10.2147/DDDT.S89621. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26491260 Free PMC article.
-
Emerging role of plumbagin: Cytotoxic potential and pharmaceutical relevance towards cancer therapy.Food Chem Toxicol. 2019 Mar;125:566-582. doi: 10.1016/j.fct.2019.01.018. Epub 2019 Jan 24. Food Chem Toxicol. 2019. PMID: 30685472 Review.
-
Pharmacological Features and Therapeutic Implications of Plumbagin in Cancer and Metabolic Disorders: A Narrative Review.Nutrients. 2024 Sep 8;16(17):3033. doi: 10.3390/nu16173033. Nutrients. 2024. PMID: 39275349 Free PMC article. Review.
Cited by
-
A SILAC-based proteomics elicits the molecular interactome of alisertib (MLN8237) in human erythroleukemia K562 cells.Am J Transl Res. 2015 Nov 15;7(11):2442-61. eCollection 2015. Am J Transl Res. 2015. PMID: 26807190 Free PMC article.
-
Estimation of the binding modes with important human cytochrome P450 enzymes, drug interaction potential, pharmacokinetics, and hepatotoxicity of ginger components using molecular docking, computational, and pharmacokinetic modeling studies.Drug Des Devel Ther. 2015 Feb 16;9:841-66. doi: 10.2147/DDDT.S74669. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 25733806 Free PMC article.
-
Expression Signatures of Long Noncoding RNAs in Left Ventricular Noncompaction.Front Cardiovasc Med. 2021 Nov 10;8:763858. doi: 10.3389/fcvm.2021.763858. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34859074 Free PMC article.
-
Total triterpenoids from Ganoderma Lucidum suppresses prostate cancer cell growth by inducing growth arrest and apoptosis.J Huazhong Univ Sci Technolog Med Sci. 2015 Oct;35(5):736-741. doi: 10.1007/s11596-015-1499-x. Epub 2015 Oct 22. J Huazhong Univ Sci Technolog Med Sci. 2015. PMID: 26489631
-
Alisertib induces G2/M arrest, apoptosis, and autophagy via PI3K/Akt/mTOR- and p38 MAPK-mediated pathways in human glioblastoma cells.Am J Transl Res. 2017 Mar 15;9(3):845-873. eCollection 2017. Am J Transl Res. 2017. PMID: 28386317 Free PMC article.
References
-
- Ahmed HU. Prostate cancer: Time for active surveillance of intermediate-risk disease? Nat Rev Urol. 2013;10(1):6–8. - PubMed
-
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v.10, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. [homepage on the Internet] Lyon, France: International Agency for Research on Cancer; 2013. [accessed on November 7, 2014]. Available from: http://globocan.iarc.fr.
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. - PubMed
-
- Soerjomataram I, Lortet-Tieulent J, Parkin DM, et al. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet. 2012;380(9856):1840–1850. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

