A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control
- PMID: 25609815
- PMCID: PMC4403431
- DOI: 10.1128/JVI.03060-14
A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control
Erratum in
-
Erratum for Saha et al., A trans-Dominant Form of Gag Restricts Ty1 Retrotransposition and Mediates Copy Number Control.J Virol. 2016 Apr 29;90(10):5210. doi: 10.1128/JVI.00418-16. Print 2016 May 15. J Virol. 2016. PMID: 27130940 Free PMC article. No abstract available.
Abstract
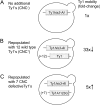
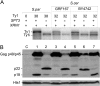
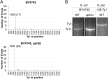
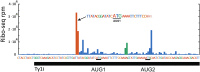
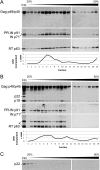
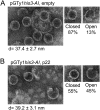
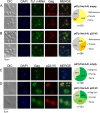
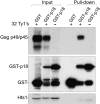
Saccharomyces cerevisiae and Saccharomyces paradoxus lack the conserved RNA interference pathway and utilize a novel form of copy number control (CNC) to inhibit Ty1 retrotransposition. Although noncoding transcripts have been implicated in CNC, here we present evidence that a truncated form of the Gag capsid protein (p22) or its processed form (p18) is necessary and sufficient for CNC and likely encoded by Ty1 internal transcripts. Coexpression of p22/p18 and Ty1 decreases mobility more than 30,000-fold. p22/p18 cofractionates with Ty1 virus-like particles (VLPs) and affects VLP yield, protein composition, and morphology. Although p22/p18 and Gag colocalize in the cytoplasm, p22/p18 disrupts sites used for VLP assembly. Glutathione S-transferase (GST) affinity pulldowns also suggest that p18 and Gag interact. Therefore, this intrinsic Gag-like restriction factor confers CNC by interfering with VLP assembly and function and expands the strategies used to limit retroelement propagation.
Importance: Retrotransposons dominate the chromosomal landscape in many eukaryotes, can cause mutations by insertion or genome rearrangement, and are evolutionarily related to retroviruses such as HIV. Thus, understanding factors that limit transposition and retroviral replication is fundamentally important. The present work describes a retrotransposon-encoded restriction protein derived from the capsid gene of the yeast Ty1 element that disrupts virus-like particle assembly in a dose-dependent manner. This form of copy number control acts as a molecular rheostat, allowing high levels of retrotransposition when few Ty1 elements are present and inhibiting transposition as copy number increases. Thus, yeast and Ty1 have coevolved a form of copy number control that is beneficial to both "host and parasite." To our knowledge, this is the first Gag-like retrotransposon restriction factor described in the literature and expands the ways in which restriction proteins modulate retroelement replication.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
The Ty1 Retrotransposon Restriction Factor p22 Targets Gag.PLoS Genet. 2015 Oct 9;11(10):e1005571. doi: 10.1371/journal.pgen.1005571. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26451601 Free PMC article.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
-
Ty1 escapes restriction by the self-encoded factor p22 through mutations in capsid.Mob Genet Elements. 2016 Mar 7;6(2):e1154639. doi: 10.1080/2159256X.2016.1154639. eCollection 2016 Mar-Apr. Mob Genet Elements. 2016. PMID: 27141327 Free PMC article.
-
Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
Cited by
-
Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA.RNA Biol. 2019 Dec;16(12):1749-1763. doi: 10.1080/15476286.2019.1657370. Epub 2019 Aug 30. RNA Biol. 2019. PMID: 31469343 Free PMC article.
-
Host-transposon interactions: conflict, cooperation, and cooption.Genes Dev. 2019 Sep 1;33(17-18):1098-1116. doi: 10.1101/gad.327312.119. Genes Dev. 2019. PMID: 31481535 Free PMC article. Review.
-
Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control.Nat Commun. 2021 Sep 22;12(1):5590. doi: 10.1038/s41467-021-25849-0. Nat Commun. 2021. PMID: 34552077 Free PMC article.
-
Specificities and Dynamics of Transposable Elements in Land Plants.Biology (Basel). 2022 Mar 23;11(4):488. doi: 10.3390/biology11040488. Biology (Basel). 2022. PMID: 35453688 Free PMC article. Review.
-
Ten things you should know about transposable elements.Genome Biol. 2018 Nov 19;19(1):199. doi: 10.1186/s13059-018-1577-z. Genome Biol. 2018. PMID: 30454069 Free PMC article. Review.
References
-
- Voytas DF, Boeke JD. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p 614–630 InCraig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

